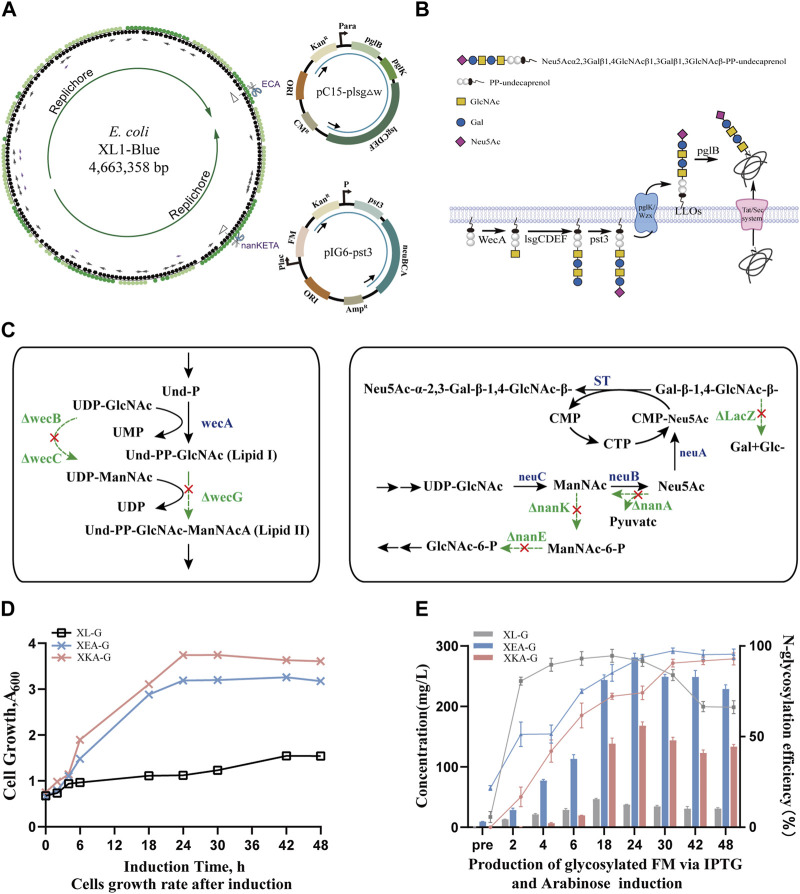
FIGURE 1.
Construction and production of human-like glyco-engineered E. coli strains. (A) A circular diagram of the E. coli XL1-Blue strain chromosome depicts the genomic integrations at ECA and nanKETA sites, alongside their corresponding replacement fragments in plasmids pC15-plsgΔw and pIG6-pst3. Plasmid pC15-plsgΔw contains the genes of oligosaccharide transferase and the required enzymes for tetrasaccharide biosynthesis; dual-expression cassette plasmid pIG6-pst3 contains genes encoding sialyltransferase and the enzymes of CMP-sialic acid biosynthesis. (B) A schematic of the biosynthesis pathway for N-glycoprotein with terminal sialylation in vivo. The O-antigen synthetase (WecA) from E. coli, glycosyltransferases (LsgCDEF) from H. influenzae, and PglK and oligosaccharide transferase (PglB) from C. jejuni are combined with sialyltransferase (Pst3) to transfer oligosaccharides to target proteins in the periplasm. (C) Metabolic pathways of knockout sites in the genome. On the left is the competing pathway of the ECA locus, on the right is the CMP-sialic acid synthesis pathway, and the bypass pathway of the nanKETA locus. (D) Cell growth curves of the glyco-engineered strain expressing glycosylated recombinant proteins modified with human-like Gal-β-1,4-GlcNAc-β-1,3-Gal-β-1,3-GlcNAc tetrasaccharide glycan under the control of arabinose promoter. (E) Production of human-like glycosylated protein using IPTG and arabinose induction. The bar graph represents the total glycoprotein yield, and the line graph illustrates the glycosylation efficiency at different time points, with the positive control protein yield being 58.5 mg/L.