Abstract
β-glucosidase hydrolyses the glycosidic bonds in cellobiose and cello-oligosaccharides, a critical step in the saccharification for biofuel production. Hence, the aim of this study was to gain insights into the biochemical and structural properties of a β-glucosidase from Beauveria bassiana, an entomopathogenic fungus. The β-glucosidase was purified to homogeneity using salt precipitation, ultrafiltration, and chromatographic techniques, attaining a specific activity of 496 U/mg. The molecular mass of the enzyme was then estimated via SDS-PAGE to be 116 kDa, while its activity pattern was confirmed by zymography using 4-methylumbelliferyl-β-d-glucopyranoside. Furthermore, the pH optima and temperature of the enzyme were found to be pH 5.0 and 60 °C respectively; its activity was significantly enhanced by Mg2+ and Na+ and was found to be relatively moderate in the presence of ethanol and dichloromethane. Molecular docking of the modelled B. bassiana β-glucosidase structure with the substrates, viz., 4-nitrophenyl β-d-glucopyranoside and cellobiose, revealed the binding affinity energies of −7.2 and −6.2 (kcal mol−1), respectively. Furthermore, the computational study predicted Lys-657, Asp-658, and Arg-1000 as the core amino acid residues in the catalytic site of the enzyme. This is the first investigation into a purified β-glucosidase from B. bassiana, providing valuable insights into the functional properties of carbohydrases from entomopathogenic fungal endophytes.
Keywords: β-glucosidase, Beauveria bassiana, Enzyme purification, In silico, Molecular docking, Structural elucidation
Highlights
-
•
First β-glucosidase from an entomopathogenic fungal endophyte purified to homogeneity.
-
•
Thermotolerant and acidophilic β-glucosidase from B. bassiana with immense potential.
-
•
Molecular docking of modelled B. bassiana β-glucosidase 3D structure with its substrates.
1. Introduction
β-glucosidase (BGL) [E.C.3.2.1.21] is a vital component of the cellulase enzyme system that hydrolyses glycosidic bonds in cellobiose and cello-oligosaccharides into the monomeric unit, glucose. The enzyme is widely distributed across nature, serving various biological functions in animals, bacteria, fungi, and plants. In bacteria and fungi, BGL catalyses the degradation of polysaccharides for energy and plant cell wall metabolism while also playing important roles in their pathogenic and symbiotic associations [1]. In plants, however, BGL is vital in cellular growth, signalling and metabolism; intermediate lignification and phytohormone activation; as well as defence against pathogens and other microbial interactions [2]. Thus, BGLs act on a wide variety of substrates, hence, they are classified based on their substrate specificity into aryl BGLs, cellobiosases, and wide range BGLs [3]. BGLs are noted to be indispensable in various industries, especially in the energy industry during cellulosic biofuel production, as well as in the food industry during winemaking and biosynthesis of flavonoid aglycones from glycosides [4].
With the global shift towards cleaner energy production, as highlighted in the United Nations Sustainable Developmental Goal 7, BGLs have become one of the most sought-after enzymes as they catalyse the rate-limiting step of cellulose hydrolysis [2]. It has been established that the efficient saccharification of cellulose is unachievable without the action of BGLs (which also removes the feedback inhibition of cellobiose), in synergy with endoglucanase and exoglucanases [1]. In this regard, there is a continuous search for BGLs with more robust properties that would enhance their applicability in the relevant bioprocesses. These desired properties include but are not limited to thermostability, pH stability, and stability in the presence of various chemical additives and solvent systems. To this end, BGLs from different microbial species, especially fungi, such as Aspergillus [5], Penicillium [6] and Fusarium species [7], have been evaluated for their potential in different industries. However, for an efficient enzymatic system, there has to be a balance between the safety of the source-organism, the functional characteristics of the synthesised enzymes as well as the cost of enzyme production.
Recent findings have shown that a strain of Beauveria bassiana, an entomopathogenic fungal endophyte, can secrete several cellulose/hemicellulose degrading enzymes including BGL, in high quantities while utilising different agricultural waste biomass as its growth substrate [8]. In addition, B. bassiana, has also been shown to be non-pathogenic to humans, as well as to plants and animals [9]. In this study, an extracellular BGL produced by B. bassiana SAN01 under submerged fermentation conditions was purified to homogeneity via precipitation, ultrafiltration, and chromatography and subjected to biochemical characterisation. Furthermore, due to the paucity of data on the structure of glycosyl hydrolases from B. bassiana, in silico approach was employed in this study to gain some valuable insights into the enzyme structure. Computational structural analyses have been established to be inexpensive, offering insightful information about the interactions, structure, and function of proteins as well as the various biological processes in which they are involved [10]. In this regard, the primary, secondary, and tertiary structures of BGL were studied to predict the possible functions associated with the enzyme active site, which may also serve as the basis for future work such as genetic manipulation, enzyme crystallography, and protein engineering. According to available literature, this is the first study aimed at identifying the functional properties of a homogenously purified β-glucosidase from an entomopathogenic fungal endophyte for future industrial applications.
2. Methodology
2.1. Materials
All the chemicals and reagents used in this study were of analytical grade and procured from Sigma Aldrich, USA. The culture media was purchased from Thermo Fisher Scientific Inc., USA while the lignocellulosic substrate, Bambara haulm, was sourced locally from Durban, South Africa.
2.2. B. bassiana SAN01 culture conditions
Beauveria bassiana SAN01 (Gene Accession Number: MN544934), was obtained from the culture collection of the Department of Biotechnology and Food Science, Durban University of Technology, South Africa. The strain was selected based on its ability to utilise readily available agricultural residues for its growth as well as its remarkable enzyme production ability. It was initially cultured on potato dextrose agar (PDA) at 30 °C for five days, and the spore suspension (1 × 106 spores mL−1) was prepared following the protocol of Amobonye, Bhagwat, Singh and Pillai [11].
2.3. β-glucosidase production
The production of B. bassiana SAN01 BGL was carried out in 250 mL Erlenmeyer flasks containing 40 gL-1 Bambara haulm as a sole of carbon source prepared in 100 mL mineral salt solution (gL−1; (0.5) MgSO4.7H2O, (0.1) FeSO4, (0. 3) K2HPO4 and (0.5) KH2PO4). The initial pH of the media was adjusted to pH 6.0 and autoclaved at 121 °C for 20 min. The media was then inoculated with the spore suspension (1 mL) and incubated at 30 °C for 9 days at 150 rpm. Subsequently, the culture broth was filtered using a sterile muslin cloth and centrifuged at 10 000×g for 15 min at 4 °C. The supernatant obtained was used as the crude enzyme for subsequent experiments.
2.4. β-glucosidase activity assay and protein estimation
β-d-glucopyranoside (pNPG) was used as the chromogenic substrate for the quantification of BGL activity. The reaction mixture comprised 0.9 mL of pNPG (5 mM) and 0.1 mL of the appropriately diluted enzyme, which was incubated at 35 °C for 30 min. The reaction was terminated by adding 1 mL of 0.5 M Na2CO3 and the absorbance was measured at 410 nm using a spectrophotometer (Shimadzu UV- 1900i). One unit of BGL activity was defined as the amount of enzyme that generated 1 μmol of p-nitrophenol in 1 min at 35 °C [6]. The protein content of the samples was determined by a modified Lowry method using bovine serum albumin as the standard [12].
2.5. β-glucosidase purification
The crude BGL was purified to homogeneity by ammonium sulphate precipitation, ultrafiltration, and gel filtration chromatography. Firstly, the crude enzyme preparation was fractionated by ammonium sulphate precipitation (60–90%) saturation. The precipitates obtained after centrifugation at 7000×g for 20 min at 4 °C were dissolved in a minimal volume of 0.1 M sodium acetate buffer (pH 5.0) and dialysed against the same buffer for 24 h at 4 °C. Subsequently, the dialysed enzyme was concentrated using a 10 kDa Amicon cut-off filter. The concentrated fraction was applied (0.5 mL) onto a Superdex 200 10/300 GL column (1.0 cm × 30 cm), equilibrated in 20 mM sodium acetate buffer (pH 5.0) and eluted with the same buffer at 0.5 mL min −1 [6] in an AKTA protein purification system (GE Healthcare Life Sciences). Subsequently, fractions were collected and assayed for BGL activity and protein content.
2.6. SDS-PAGE and zymogram
The molecular weight of the purified BGL was estimated using 12% cross-linked polyacrylamide gel and 4% stacking gel. Enzyme aliquots of 10 μg were loaded into the sample wells and electrophoresed at a constant voltage of 100 V for 2 h at room temperature. After electrophoresis, the gel was stained with silver staining [13]. Subsequently, zymography was conducted using a native gel electrophoresis with 7% polyacrylamide gel and 4% stacking gel. After electrophoresis, the gel was immersed in 4-methylumbelliferyl-β-d-glucopyranoside prepared in 0.1 mM sodium acetate buffer, pH 5.0 at 45 °C for 30 min in the dark. The release of methylumbelliferone from the substrate was observed under UV at 310 nm [14].
2.7. Biochemical characterisation of β-glucosidase
2.7.1. pH optima and pH stability
To determine the optimum pH for BGL, different buffer preparations including acetate (pH 3.0–5.0), phosphate (pH 6.0–7.0), and Tris-HCl (pH 8.0–9.0) at 0.05 M were used. The enzyme was pre-incubated in the respective buffer for 30–300 min at 35 °C for the stability test and the residual activities were measured accordingly [15].
2.7.2. Temperature optima and thermostability
The optimum temperature for BGL was evaluated by incubating the reaction mixtures between 25 and 70 °C at 5 °C intervals. Subsequently, the thermostability of BGL was evaluated by pre-incubating the enzyme between 30 and 50 °C for 300 min; samples were taken at 60 min intervals [16]. Relative activity was measured according to standard enzyme assay [16].
2.7.3. Effect of organic solvents on enzyme activity
The inhibitory or stimulatory effects of the different organic solvents, viz., acetone, benzene, ethanol, butanol, hexane, methanol, and toluene on BGL activity were assessed. The enzyme was incubated for 1 h at room temperature in solutions containing each solvent (10% v/v) with continuous agitation at 120 rpm [17]. Subsequently, aliquots were removed, and the relative activity was measured according to the standard enzyme assay mentioned in section 2.4.
2.7.4. Effect of metal ions and salt concentration on enzyme activity
The effects of selected metal ions including Ba2+ (BaCl2), Co2+ (CoCl2), Fe2+ (FeSO4), Mg2+ (MgSO4), Cu2+ (CuSO4), Zn2+ (ZnSO4), Na+ (NaCl) and Hg2+ (HgCl2) on the BGL activity were evaluated at final concentrations of 1 mM and 10 mM [18].
2.7.5. Effect of various additives on enzyme activity
The effects of selected additives such as β-mercaptoethanol (BME), dithiothreitol (DTT), ethylenediaminetetraacetic acid (EDTA), phenylmethylsulphonyl fluoride (PMSF), sodium dodecyl sulphate (SDS), Triton X-100 and Tween 20 on the BGL activity were evaluated using 1 mM and 10 mM of the additive [19]. All residual activities were measured under standard enzyme assay conditions.
2.8. In silico structural characterisation of B. bassiana β-glucosidase
2.8.1. Sequence retrieval and primary analysis
The sequence of B. bassiana SAN01 BGL was selected from the NCBI database (Accession no: KAH8714014.1) based on the estimated molecular weight of the purified enzyme in this study. The ProtParam tool was used to compute the aliphatic index, isoelectric point, the total number of negatively and positively charged residues, the instability index, and the grand average of hydropathicity. Subsequently, PSIPRED server http://bioinf.cs.ucl.ac.uk/psipred/), and Pfam search (http://pfam.xfam.org/) were employed in predicting the secondary structure and domains present in the BGL, respectively [20].
2.8.2. Homology modelling and molecular docking
The 3D structure of B. bassiana SAN01 BGL was modelled using the template with the highest sequence identity and coverage. It was selected for homology modelling, subsequently; the quality of the predicted structure was evaluated by the Ramachandran plot which highlighted the energetically allowed regions [10].
2.8.3. Molecular docking
Initially, the active sites of the BGL were predicted using CASTp and MetaPocket 2.0 [21,22]. Subsequently, Autodock 4.2 software was used for docking the modelled 3D structure of B. bassiana SAN01 BGL as the target and its two substrates, cellobiose, and p-nitrophenyl β-d-glucopyranoside, as the ligands. The structures of the ligands -cellobiose (PubChem CID: 294), and p-nitrophenyl β-d-glucopyranoside (PubChem CID: 92930) were obtained from the PubChem database (http://pubchem.ncb.nlm.nih.gov/) in SDF format and converted into PDB format using BIOVA Discovery Studio Visualizer (BIOVA, CA, USA). Polar hydrogens were added to the target molecule using Autodock tools, and a 40 × 40 × 40 grid box was used in the configuration file of Autodock Vina with the box centred at X: 32.128, Y: 26.721, Z: 27.453 coordinates. The receptor atom positions were fixed, and the torsion angle of the ligand glycosidic bonds was rotated to attain favourable docking. All other docking parameters were set to default. Subsequently, the most suitable BGL-ligand pose was chosen and visualised using PyMol [23].
2.9. Statistical analyses
Statistical variance (ANOVA) was calculated with GraphPad Prism software (version 10) and data were all presented as the mean ± standard deviation of triplicate values. Differences between samples were undertaken to be statistically significant if p < 0.05.
3. Results and discussion
3.1. Purification of B. bassiana β-glucosidase
After nine days of submerged fermentation, a BGL activity of 148.86 U/mL was recorded from the clarified crude fermentation broth of B. bassiana SAN01. The production level of BGL by B. bassiana SAN01 obtained in this study is the highest reported to date for any entomopathogenic fungus, and it is significantly higher when compared to the previously reported 2.5 U/mL from another strain of B. bassiana [24]. Subsequently, B. bassiana SAN01 BGL was purified to homogeneity, as summarised in Table 1. The purification approach resulted in a homogenous enzyme with a specific activity of 496 U/mg, which was higher than some previously reported BGLs such as those from A. niger [5] and Penicillium citrinum UFV1 [6] which had specific activities of 60.6 U/mg, and 349.2 U/mg, respectively.
Table 1.
Purification table of B. bassiana SAN01 β-glucosidase.
| Purification steps | Total activity (U) | Total protein (mg)a | Specific activity | Recovery (%) |
|---|---|---|---|---|
| Crude enzyme | 14886 ± 483 | 383 ± 18.7 | 38.9 | 100 |
| NH4(SO4)2 precipitation | 5532 ± 104 | 73 ± 2.7 | 75.8 | 37.2 |
| Ultrafiltration | 2685 ± 61 | 28 ± 1.3 | 95.9 | 18 |
| Gel filtration chromatography | 1482 ± 26 | 2.99 ± 0.07 | 496 | 10 |
Total protein content measured by Lowry-Hartree assay using BSA as the standard.
The analysis of the protein pattern of the purified B. bassiana BGL revealed a single band (Fig. 1a) with an estimated size of 116 kDa using SDS-PAGE. The size of the protein was also confirmed via zymography, which showed the band at a similar position as observed on SDS-PAGE (Fig. 1b). In the zymogram, it was observed that the BGL hydrolysed 4-methylumbelliferyl-β-d-glucopyranoside to release methylumbelliferone which fluoresced under UV light. The molecular mass obtained for B. bassiana SAN01 BGL was close to those recorded for the same enzyme from some Aspergillus spp. such as A. versicolor and A. fumigatus JCM 10253 with molecular mass of 100 kDa [16] and 125 kDa, respectively [17]. However, BGL with a similar molecular mass was also obtained by Pal [25] from Termitomyces clypeatus, another filamentous fungus.
Fig. 1.
(a)B. bassiana SAN01 β-glucosidase on 12% SDS-PAGE- Lane 1: Purified enzyme, Lane 2: Ammonium sulphate fraction, Lane 3: Crude enzyme, M: wide range protein marker (14–250 kDa). (b) B. bassiana β-glucosidase activity on 7% Native PAGE gel containing 50 mM 4-methylumbelliferyl β-D-glucoside - AF: active purified fraction, M: wide range protein marker (14–250 kDa). Original images of the SDS-PAGE, zymogram and wide range marker are provided in Supplementary data as figure S1, figure S2 and figure S3 respectively.
3.2. Characterisation of B. bassiana SAN01 β-glucosidase
3.2.1. Effect of pH on B. bassiana SAN01 β-glucosidase
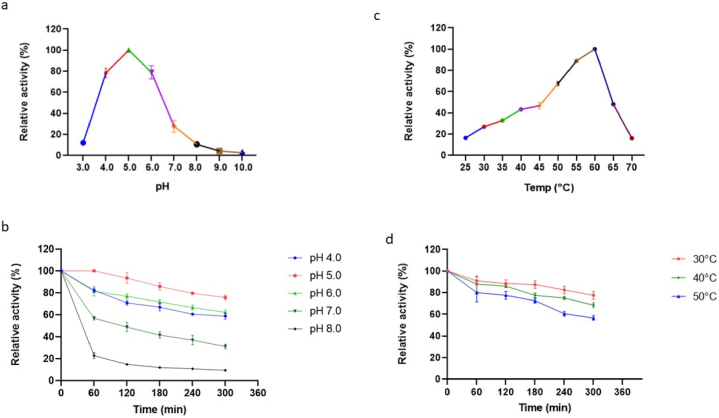
The optimum pH for B. bassiana BGL was observed to be at pH 5.0 while substantial enzyme activity (83%) was also observed at pH 6.0. However, a further increase in pH led to a decline in the activity, suggesting that the BGL from B. bassiana SAN01 is a moderately acidophilic enzyme (Fig. 2a). Concurring with these results, unpurified BGL from a different B. bassiana strain was previously shown to be active under moderately acidic conditions [24], in addition, most fungal BGLs have been reported to be active at pH 4.0–6.0. Recent examples include the BGL from Aspergillus fumigatus [26] and Neofusicoccum parvum [27]. The BGL in this study retained >60 residual activity after 4 h incubation at pH 4.0, 5.0, and 6.0, respectively (Fig. 2b). It was also observed that the enzyme retained more than 80% of its initial activity at its optimum pH of 5.0 after 4 h while also retaining more than 65% of activity at pHs 4.0 and 6.0. Thus, B. bassiana SAN01 BGL demonstrates a wide range of stability at acidic pH, which makes it suitable for use in industrial bioprocesses that require acidic conditions. However, at pH values above 7.0 the enzyme activity was observed to drop, indicating the low tolerance of the enzyme to alkaline conditions.
Fig. 2.
(a) Effect of pH on B. bassiana β-glucosidase activity; (b) Effect of pH on the stability of B. bassiana β-glucosidase (c) Effect of temperature on B. bassiana β-glucosidase activity (d) Effect of temperature on the stability of B. bassiana β-glucosidase (Each point represents the mean (n = 3) ± SD; enzyme activities in a & c were calculated relative to the highest activity recorded; enzyme activities in b & d were calculated relative to the highest activity recorded at t = 0).
3.2.2. Effect of temperature on B. bassiana SAN01 β-glucosidase
B. bassiana SAN01 BGL displayed maximal activity at 60 °C among the different temperatures evaluated, which is typical of a thermophilic microorganism (Fig. 2c). Borgi [24] had previously recorded an optimal temperature of 55 °C for crude BGL preparation from another B. bassiana strain. Similarly, other mesophilic fungi besides B. bassiana have been recorded to produce BGL with optimum activity between 40 and 60C°, such as Aspergillus flavus [28], A. fumigatus JCM 10253 [17], and Aspergillus sp. DHE7 [29]. Furthermore, it was observed that the B. bassiana BGL retained >50% of its initial activity after 300 min of incubation between 30 °C and 50 °C, however, its stability declined above 50 °C. The enzyme was also observed to retain ∼85% of its activity at 30 °C followed by ∼75% at 40 °C (Fig. 2d). In this regard, B. bassiana SAN01 BGL could be suitable for applications carried out at mid-temperature conditions, however, B. bassiana SAN01 BGL exhibited higher stability profile when compared to recombinant BGL from Alteromonas sp. which exhibited a t1/2 of 20 min at 40 °C [30], and Microbulbifer sp. ALW1 BGL which showed less than 20% of the original activity after 120 min at a temperature between 40 and 45 °C [31].
3.3. Effect of metal ions, solvents, and additives on B. bassiana β-glucosidase activity
Enzyme activity could significantly be altered in the presence of different metal ions such as Co2+, Fe2+, Mg2+, Ag2+, Cu2+, and Hg2+; hence, it is essential to evaluate the enzyme activity in the presence of metal ions to identify their activating or inhibitory potential. The metal ions selected in this study have been shown in previous studies to exhibit varying degrees of interactions with enzymes, including BGLs, with some of them acting as activators, co-factors, inhibitors, modulators as well as contributing to the tertiary structural stability of enzymes [32]. In this study, a significant portion of the BGL activity was maintained in the presence of all the metal ions evaluated at a concentration of 1 mM, while the activity was significantly enhanced by Mg2+ followed by Co2+ and Na+ (Table 2). However, at an increased ionic concentration of 10 mM, the initial activity was only marginally enhanced by Na+ and Mg2+ suggesting that these two metal ions could be co-factors for B. bassiana SAN01 BGL; the activity was observed to be maintained to a significant degree by Ag2+ Cu2+, and Zn2+. However, the activity of the BGL was found to be almost completely inhibited by the higher concentration of Hg2+ (Table 2). The positive effect of Mg2+, Co2+, and Na + on the activity of BGL from other organisms such as Myceliophthora thermophila M.7.7 [15], and Penicillium citrinum UFV1 was previously reported. Hg2+ on the other hand has been noted to inhibit the enzyme activity in most cases [6,30] and the inhibitory effect could be via its interaction with cysteine residues on sulfhydryl groups, an interaction that modifies the tertiary structure of the protein leading to denaturation [17].
Table 2.
Effect of metal ions on B. bassiana SAN01 β-glucosidase.
| Metal ion | Relative activity (%) |
|
|---|---|---|
| 1 mM | 10 mM | |
| Control | 100 | 100 |
| Ag2+ | 88.8 ± 4.7 | 67.3 ± 3.7 |
| Co2+ | 121.6 ± 6.7 | 100.2 ± 4.9 |
| Cu2+ | 94.1 ± 5.7 | 88.8 ± 4.1 |
| Fe2+ | 104.6 ± 6.4 | 57.9 ± 2.4 |
| Hg2+ | 95.4 ± 4.3 | 3.9 ± 3.4 |
| Mg2+ | 168.1 ± 8.5 | 110.5 ± 5.3 |
| Na+ | 117.8 ± 6.1 | 110.5 ± 4.9 |
| Zn2+ | 98.4 ± 5.5 | 88.6 ± 4.6 |
Data are shown as mean ± SD (n = 3).
aNo chemical added to control.
3.3.1. Effect of additives on B. bassiana SAN01 β-glucosidase
Different additives and surfactants have varying effects on hydrolytic enzymes, especially during industrial applications. As there is an inexhaustive list of additives, the selected ones were based on previous literature and their perceived influence on the industrial applicability of the enzyme. For example, the detergents- Triton-X-100, Tween 80, and Tween 20- were studied due to the potential use of cellulases in detergent formulation [33]. Furthermore, the reaction of some additives with an enzyme could also give some insights into the structure and biology of the enzyme. For instance, DTT is known to disrupt disulphide bonds in proteins, hence, the intensity of its stimulatory or inhibitory activity is linked to the relevance of the bonds in the enzyme's structure and activity [34]. In this study, none of the tested additives and surfactants stimulated B. bassiana SAN01 BGL activity, however, the enzyme was observed to be marginally stable in DTT, EDTA, PMSF, Triton-X-100, Tween 80, and Tween 20 at both concentrations assessed (Table 3). On the other hand, the enzyme activity was significantly inhibited by SDS at both 1 mM and 10 mM concentrations. The inhibitory effect of SDS was previously reported on BGL from A. flavus [28], Microbulbifer sp. ALW1 [31], and Penicillium roqueforti [35]. The negative effects of SDS on enzyme activity have been ascribed to the detergent's interference with the enzymes' hydrophobic regions as well as the tertiary structure, leading to denaturation [20].
Table 3.
Effect of additives on B. bassiana SAN01 β-glucosidase.
| Additives | Relative activity (%) |
|
|---|---|---|
| 1 mM | 10 mM | |
| Control | 100 | 100 |
| β-mercaptoethanol | 76.9 ± 2.2 | 66.1 ± 0 |
| DTT | 86.9 ± 0.4 | 59.8 ± 0.5 |
| EDTA | 85.5 ± 0.4 | 74.8 ± 0.4 |
| PMSF | 85.7 ± 1.8 | 70.1 ± 1.4 |
| SDS | 21.6 ± 0.6 | 3.1 ± 0.2 |
| Triton -X 100 | 95.2 ± 1.8 | 79.8 ± 0.2 |
| Tween 20 | 80.9 ± 5.1 | 68.74 ± 0.46 |
| Tween 80 | 80.7 ± 1.4 | 76.5 ± 2.6 |
Data are shown as mean ± SD (n = 3).
a No solvent was added in control.
3.3.2. Effect of organic solvents on B. bassiana SAN01 β-glucosidase
The industrial applications of enzymes involve the presence of several organic solvents. In some cases, these solvents may form a significant portion or the whole final products of the reaction, or they might be required for the dissolution of the substrates or intermediate products. Various organic solvents have also been found useful during protein purification, especially as precipitating agents. Thus, the organic solvents were selected in this study based on their industrial importance as well as on the basis of previous reports [24,36,33].
The effect of the selected organic solvents on B. bassiana SAN01 BGL activity was evaluated by determining the residual activity of the enzyme in the presence of each solvent. B. bassiana SAN01 BGL was observed to retain 80% and 71% of its activity in the presence of dichloromethane and ethanol, respectively (Table 4). In contrast, its activity was strongly inhibited in the presence of acetone, butanol, chloroform, hexane, and isopropanol at the concentrations tested (Table 4). The stability of fungal BGL in more polar organic solvents such as ethanol has been previously reported from A. niger [5], and Dictyoglomus turgidum [37]. Biocatalytic reactions that are carried out in the organic phase have been reported to offer several advantages such as easy recyclability, lower microbial contamination by undesired organisms, and increased solubility of the hydrophobic substrate, facilitating effective reactions [38]. Hence, our results suggest that B. bassiana SAN01 BGL could be channelled into industrial applications that involve the use of organic solvents; for example, B. bassiana SAN01 BGL could be utilised in one-pot synthesis of ethanol which involves the coupling of the saccharification and alcohol fermentation in the same vessel.
Table 4.
Effect of organic solvents on the activity B. bassiana SAN01 β-glucosidase.
| Solvents | Relative activity (%) |
|---|---|
| Control | 100 |
| Acetone | 9.9 ± 0.3 |
| Butanol | 5.2 ± 0.3 |
| Chloroform | 2 ± 0.1 |
| Dichloromethane | 81.4 ± 3.4 |
| Ethanol | 70.1 ± 3.5 |
| Hexane | 5.9 ± 0.2 |
| Isopropanol | 3.3 ± 0.1 |
Data are shown as mean ± SD (n = 3).
No chemical added to control.
3.3.3. In silico structural prediction of B. bassiana SAN01 β-glucosidase
According to available literature, there is no information on the secondary and tertiary structure of any lignocellulolytic enzyme from B. bassiana SAN01 including BGLs, hence it is considered imperative to gain a preliminary insight into the structure of BGL using a computational approach. Subsequent to the estimation of the molecular weight of the purified BGL via SDS-PAGE, a sequence was selected from ∼30 B. bassiana BGL sequences in the NCBI database for further analysis. The BGL sequence (Accession no: KAH8714014.1) was selected based on its approximated molecular weight of 116 kDa, which is close to the one obtained in this study. Analysis of the primary sequence showed that the enzyme comprises 1044 amino acids; in addition, the enzyme was predicted to have a molecular weight and isoelectric point (pI) of ∼116 kDa and 5.59, respectively. The enzyme was also observed to contain more negatively charged amino acid residues (aspartic acid and glutamic acid = 116) than positive residues (arginine and lysine = 97). The protein was also predicted to be quite stable judging from its computed instability index of 34.40, thus corroborating the results on the thermostability and pH stability obtained earlier in this study.
The B. bassiana BGL sequence was predicted with an aliphatic index (AI) and Grand average of hydropathicity (GRAVY) of 71.98 and −0.399, respectively. AI has been used to predict protein thermostability based on the fraction of aliphatic amino acids in the protein structure, and higher AI values are synonymous with higher thermostability [39,40]. Thus, the AI value of 71.78 obtained in this study, provides additional validation to notable thermostability obtained during characterisation studies. According to previous studies, negative GRAVY values may indicate the hydrophilicity of a protein [20,41]. Thus, from the results, B. bassiana SAN01 BGL can be predicted to be hydrophilic overall, raising the probability of its significant stability in an aqueous environment. It was also observed from the domain analysis that B. bassiana SAN01 BGL belongs to the glycosyl hydrolases 1 family which also encompasses other accessory enzymes such as β-mannosidase, β-d-fucosidase, β-glucuronidase and 6-phospho- β-galactosidase.
For homology modelling of the B. bassiana SAN01 BGL, the alpha model structure of B. bassiana ARSEF 2860 (PDB ID: J5JSG7_BEAB2) was chosen as the template, out of the 12 templates that were generated, as it had the highest sequence identity (96.68%) and coverage (0.98) of all the suggested templates. The visualisation of the model revealed that the generated model is built on the barrel type (β/α)8 architecture, forming coin slot-like, deep, and narrow active site (Fig. 3a). This structure is conserved in the GH family 1 and has been observed in glucosidases from Phanerochaete chrysosporium [42] and Coniophora puteana [43]. Furthermore, the Ramachandran plot of the B. bassiana BGL showed that the total distribution of amino acid residues in the favoured region is 92.7%, which reflects acceptable model adequacy, while a significantly lower percentage of the residues were found in the outlier region (0.86 %) (Fig. 3b). Furthermore, the overall quality factor score predicted for the BGL model by ERRAT was 92.33 while VERIFY 3D also showed that 95.69% of the amino acid residues have an average 3D-ID score of ≥ 0.1, thus, pointing to the reliability of the generated model.
Fig. 3.
(a) 3D structure of B. bassiana SAN01 BGL (b) Ramachandran plot of B. bassiana SAN01 BGL; 92.71% favoured region, 0.86% outlier.
The docked complex of B. bassiana SAN01 BGL with the ligands showed the binding affinity scores of −6.2 and −7.2 (kcal mol−1) for cellobiose and pNPG, respectively, thus, indicating that B. bassiana BGL is better orientated to the artificial substrate pNPG than the natural substrate, cellobiose. In accordance, a similar binding affinity score of −6.2 for cellobiose was obtained by Khairudin and Mazlan [44] with Paenobacillus polymyxa BGL. The analysis of the pNPG-BGL complex revealed that the ligand interacted with the active site forming hydrogen bonds with the Lys 652, Lys 658, Arg 1000, and Arg 974 residues (Fig. 4). For the cellobiose-BGL complex, however, the hydrogen bond interactions were formed with Asp 658, Arg 974, and Ser 1001 (Fig. 5). All the molecular interactions recorded in both enzyme-ligand complexes are presented in Table 5. These results indicate that B. bassiana SAN01 BGL binds significantly with both natural and synthetic substrates unlike the BGL from Neosartorya fischeri which showed no hydrogen bond interaction with cellobiose [45]. Hence, the BGL in this study can be classified into the third class of glucosidase which acts on both aryl- β-D-glucosides and disaccharides due to the broad substrate specificity.
Fig. 4.
B. bassiana SAN01 BGL-pNPG complex and molecular interactions between pNPG and ligand sites of B. bassiana SAN01 BGL.
Fig. 5.
B. bassiana SAN01 BGL-cellobiose complex and interactions between cellobiose and ligand sites of B. bassiana SAN01 BGL.
Table 5.
Active site interaction in B. bassiana SAN01 BGL-substrate complexes.
| Substrate | Interactive bonds between enzyme and ligands | Active site amino acids |
|---|---|---|
| pNPG | Hydrogen | Lys 652, Lys 658, Arg 1000, Arg 974 |
| Carbon-hydrogen | Asp 658, Asp 661, Asn 999 | |
| Pi cation, Pi anion | Arg 1000, Asp 658 | |
| Adverse acceptor– acceptor | Asp 661 | |
| Hydrogen | Ser 1001, Asp 658, Arg 974 | |
| Cellobiose | Carbon-hydrogen | Lys 657, Arg 100 |
| Adverse acceptor-acceptor | Asn 999, Tyr 642 |
4. Conclusion
In this study BGL from a fungal endophyte, B. bassiana SAN01 was purified to homogeneity for the first time. This particular β-glucosidase exhibited several noteworthy characteristics, including stability at acidic pH levels, resistance to various metals, and tolerance to ethanol. Additionally, molecular studies indicate that B. bassiana SAN01 BGL exhibits activity towards both aryl-β-D-glycosides and disaccharides. Consequently, these findings imply that B. bassiana SAN01 BGL can be classified as a class Ⅲ BGL, owing to its wider range of substrate specificity. This characteristic makes it a promising candidate for various biotechnological applications, such as bioethanol production from lignocellulosic materials, food processing, and the biosynthesis of aryl glycosides in the pharmaceutical industry. However, additional investigation is necessary to examine its potential applicability further, particularly when used in conjunction with other cellulase enzymes, to develop more effective enzyme cocktails for improved saccharification of lignocellulosic biomass.
Availability of data and material
Data will be made available on request.
CRediT authorship contribution statement
Buka Magwaza: Writing – original draft, Investigation, Formal analysis. Ayodeji Amobonye: Writing – review & editing, Investigation, Formal analysis, Conceptualization, Supervision. Prashant Bhagwat: Writing – review & editing, Investigation. Santhosh Pillai: Writing – review & editing, Supervision, Resources, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by the National Research Foundation of South Africa under grant numbers [UID 146320, and UID 138097].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e28667.
Contributor Information
Buka Magwaza, Email: bukawelcome4359@gmail.com.
Ayodeji Amobonye, Email: dejiamobonye@gmail.com, ayodejia1@dut.ac.za.
Prashant Bhagwat, Email: pkbhagwat9988@gmail.com, prashantb@dut.ac.za.
Santhosh Pillai, Email: santhoshk@dut.ac.za.
Appendix ASupplementary data
The following is the Supplementary data to this article.
References
- 1.Singh G., Verma A., Kumar V. Catalytic properties, functional attributes and industrial applications of β-glucosidases. 3 Biotech. 2016;6:1–14. doi: 10.1007/s13205-015-0328-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Godse R., Bawane H., Tripathi J., Kulkarni R. Unconventional β-Glucosidases: a promising biocatalyst for industrial biotechnology. Appl. Biochem. Biotechnol. 2021;193:2993–3016. doi: 10.1007/s12010-021-03568-y. [DOI] [PubMed] [Google Scholar]
- 3.Liew K.J., Lim L., Woo H.Y., Chan K.G., Shamsir M.S., Goh K.M. Purification and characterization of a novel GH1 beta-glucosidase from Jeotgalibacillus malaysiensis. Int. J. Biol. Macromol. 2018;115:1094–1102. doi: 10.1016/j.ijbiomac.2018.04.156. [DOI] [PubMed] [Google Scholar]
- 4.Nair A., Kuwahara A., Nagase A., Yamaguchi H., Yamazaki T., Hosoya M., Omura A., Kiyomoto K., Yamaguchi M.A., Shimoyama T. Purification, gene cloning, and biochemical characterization of a β-glucosidase capable of hydrolyzing sesaminol triglucoside from Paenibacillus sp. KB0549. PLoS One. 2013;8 doi: 10.1371/journal.pone.0060538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narasimha G., Sridevi A., Ramanjaneyulu G., Rajasekhar Reddy B. Purification and characterization of β-glucosidase from Aspergillus niger. Int. J. Food Prop. 2016;19:652–661. doi: 10.1080/10942912.2015.1023398. [DOI] [Google Scholar]
- 6.da Costa S.G., Pereira O.L., Teixeira-Ferreira A., Valente R.H., de Rezende S.T., Guimarães V.M., Genta F.A. Penicillium citrinum UFV1 β-glucosidases: purification, characterization, and application for biomass saccharification. Biotechnol. Biofuels. 2018;11:1–19. doi: 10.1186/s13068-018-1226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertonha L.C., Neto M.L., Garcia J.A.A., Vieira T.F., Castoldi R., Bracht A., Peralta R.M. Screening of Fusarium sp. for xylan and cellulose hydrolyzing enzymes and perspectives for the saccharification of delignified sugarcane bagasse. Biocatal. Agric. Biotechnol. 2018;16:385–389. doi: 10.1016/j.bcab.2018.09.010. [DOI] [Google Scholar]
- 8.Amobonye A., Bhagwat P., Singh S., Pillai S. Enhanced xylanase and endoglucanase production from Beauveria bassiana SAN01, an entomopathogenic fungal endophyte. Fungal Biol. 2021;125:39–48. doi: 10.3390/jof7080668. [DOI] [PubMed] [Google Scholar]
- 9.Keswani C., Singh S.P., Singh H.B. Beauveria bassiana: status, mode of action, applications and safety issues. Biotechnol. Today. 2013;3:16–20. doi: 10.5958/j.2322-0996.3.1.002. [DOI] [Google Scholar]
- 10.Amobonye A., Singh S., Mukherjee K., Jobichen C., Qureshi I.A., Pillai S. Structural and functional insights into fungal glutaminase using a computational approach. Process Biochem. 2022;117:76–89. doi: 10.1016/j.procbio.2022.03.01. [DOI] [Google Scholar]
- 11.Amobonye A., Bhagwat P., Singh S., Pillai S. Beauveria bassiana xylanase: characterization and wastepaper deinking potential of a novel glycosyl hydrolase from an endophytic fungal entomopathogen. J. Fungi. 2021;7:668. doi: 10.3390/jof7080668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartree E. Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal. Riochem. 1972;48:422–427. doi: 10.1016/0003-2697(72)90094-2. [DOI] [PubMed] [Google Scholar]
- 13.González-Pombo P., Pérez G., Carrau F., Guisán J.M., Batista-Viera F., Brena B.M. One-step purification and characterization of an intracellular β-glucosidase from Metschnikowia pulcherrima. Biotechnol. Lett. 2008;30:1469–1475. doi: 10.1007/s10529-008-9708-3. [DOI] [PubMed] [Google Scholar]
- 14.Kuo H.P., Wang R., Huang C.Y., Lai J.T., Lo Y.C., Huang S.T. Characterization of an extracellular β-glucosidase from Dekkera bruxellensis for resveratrol production. J. Food Drug Anal. 2018;26:163–171. doi: 10.1016/j.jfda.2016.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonfá E.C., de Souza Moretti M.M., Gomes E., Bonilla-Rodriguez G.O. Biochemical characterization of an isolated 50 kDa beta-glucosidase from the thermophilic fungus Myceliophthora thermophila M. 7.7. Biocatal. Agric. Biotechnol. 2018;13:311–318. doi: 10.1016/j.bcab.2018.01.008. [DOI] [Google Scholar]
- 16.Huang C., Feng Y., Patel G., Xu X.-q., Qian J., Liu Q., Kai G.-y. Production, immobilization and characterization of beta-glucosidase for application in cellulose degradation from a novel Aspergillus versicolor. Int. J. Biol. Macromol. 2021;177:437–446. doi: 10.1016/j.ijbiomac.2021.02.154. [DOI] [PubMed] [Google Scholar]
- 17.Saroj P., Narasimhulu K. Biochemical characterization of thermostable carboxymethyl cellulase and β-glucosidase from Aspergillus fumigatus JCM 10253. Appl. Biochem. Biotechnol. 2022;194:2503–2527. doi: 10.1007/s12010-022-03839-2. [DOI] [PubMed] [Google Scholar]
- 18.Sun N., Liu X., Zhang B., Wang X., Na W., Tan Z., Li X., Guan Q. Characterization of a novel recombinant halophilic β-glucosidase of Trichoderma harzianum derived from Hainan mangrove. BMC Microbiol. 2022;22:1–11. doi: 10.1186/s12866-022-02596-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.da Silva R.R., da Conceição P.J.P., de Menezes C.L.A., de Oliveira Nascimento C.E., Bertelli M.M., Júnior A.P., de Souza G.M., da Silva R., Gomes E. Biochemical characteristics and potential application of a novel ethanol and glucose-tolerant β-glucosidase secreted by Pichia guilliermondii G1. 2. J. Biotechnol. 2019;294:73–80. doi: 10.1016/j.jbiotec.2019.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Bhagwat P., Amobonye A., Singh S., Pillai S. A comparative analysis of GH18 chitinases and their isoforms from Beauveria bassiana: an in-silico approach. Process Biochem. 2021;100:207–216. doi: 10.1016/j.procbio.2020.10.012. [DOI] [Google Scholar]
- 21.Pathaw N., Gurung A.B., Chrungoo N.K., Bhattacharjee A., Roy S.S., Ansari M.A., Sharma S.K. In silico molecular modelling, structural dynamics simulation and characterization of antifungal nature of β-glucosidase enzyme from Sechium edule. J. Biomol. Struct. Dynam. 2021;39(12):4501–4509. doi: 10.1080/07391102.2020.1791956. [DOI] [PubMed] [Google Scholar]
- 22.Singh S., Ahmed J., Gavande P.V., Fontes C.M., Goyal A. Structural and functional insights into the glycoside hydrolase family 30 xylanase of the rumen bacterium Ruminococcus flavefaciens. J. Mol. Struct. 2023;1272 doi: 10.1016/j.molstruc.2022.134155. [DOI] [Google Scholar]
- 23.Zada N.S., Belduz A.O., Güler H.I., Khan A., Sahinkaya M., Kaçıran A., Ay H., Badshah M., Shah A.A., Khan S. Cloning, expression, biochemical characterization, and molecular docking studies of a novel glucose tolerant β-glucosidase from Saccharomonospora sp. NB11. Enzym. Microb. Technol. 2021;148 doi: 10.1016/j.enzmictec.2021.109799. [DOI] [PubMed] [Google Scholar]
- 24.Borgi I., Gargouri A. A novel high molecular weight thermo-acidoactive β-glucosidase from Beauveria bassiana. J. Appl. Biochem. Microbiol. 2016;52:602–607. [Google Scholar]
- 25.Pal S., Banik S.P., Ghorai S., Chowdhury S., Khowala S. Purification and characterization of a thermostable intra-cellular β-glucosidase with transglycosylation properties from filamentous fungus Termitomyces clypeatus. Bioresour. Technol. 2010;101:2412–2420. doi: 10.1016/j.biortech.2009.11.064. [DOI] [PubMed] [Google Scholar]
- 26.Saroj P., P M., Narasimhulu K. Biochemical characterization of thermostable carboxymethyl cellulase and β-glucosidase from Aspergillus fumigatus JCM 10253. Appl. Biochem. Biotechnol. 2022;194:2503–2527. doi: 10.1007/s12010-022-03839-2. [DOI] [PubMed] [Google Scholar]
- 27.Singh N., Sithole B., Kumar A., Govinden R. A glucose tolerant β-glucosidase from a newly isolated Neofusicoccum parvum strain F7: production, purification, and characterization. Sci. Rep. 2023;13:5134. doi: 10.1038/s41598-023-32353-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Z., Liu Y., Liu L., Chen Y., Li S., Jia Y. Purification and characterization of a novel β-glucosidase from Aspergillus flavus and its application in saccharification of soybean meal. Prep. Biochem. Biotechnol. 2019;49:671–678. doi: 10.1080/10826068.2019.1599397. [DOI] [PubMed] [Google Scholar]
- 29.El-Ghonemy D.H. Optimization of extracellular ethanol-tolerant β-glucosidase production from a newly isolated Aspergillus sp. DHE7 via solid state fermentation using jojoba meal as substrate: purification and biochemical characterization for biofuel preparation. J. Genet. Eng. Biotechnol. 2021;19:1–18. doi: 10.1186/s43141-021-00144-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun J., Wang W., Yao C., Dai F., Zhu X., Liu J., Hao J. Overexpression and characterization of a novel cold-adapted and salt-tolerant GH1 β-glucosidase from the marine bacterium Alteromonas sp. L82, J. Microbiol. 2018;56:656–664. doi: 10.1007/s12275-018-8018-2. [DOI] [PubMed] [Google Scholar]
- 31.Jiang Z., Long L., Liang M., Li H., Chen Y., Zheng M., Ni H., Li Q., Zhu Y. Characterization of a glucose-stimulated β-glucosidase from Microbulbifer sp. ALW1. Microbiol. Res. 2021;251 doi: 10.1016/j.micres.2021.126840. [DOI] [PubMed] [Google Scholar]
- 32.Andreini C., Bertini I., Cavallaro G., Holliday G.L., Thornton J.M. Metal ions in biological catalysis: from enzyme databases to general principles. JBIC J. Biol. Inorgan. Chem. 2008;13:1205–1218. doi: 10.1007/s00775-008-0404-5. [DOI] [PubMed] [Google Scholar]
- 33.Biver S., Stroobants A., Portetelle D., Vandenbol M. Two promising alkaline β-glucosidases isolated by functional metagenomics from agricultural soil, including one showing high tolerance towards harsh detergents, oxidants and glucose. J. Industrial. Microbiol. Biotechnol. 2014;41(3):479–488. doi: 10.1007/s10295-014-1400-0. [DOI] [PubMed] [Google Scholar]
- 34.Fan F., Zhang Q., Zhang Y., Huang G., Liang X., Wang C.-c., Wang L., Lu D. Two protein disulfide isomerase subgroups work synergistically in catalyzing oxidative protein folding. Plant physiol. 2022;188(1):241–254. doi: 10.1093/plphys/kiab457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.das Neves C.A., de Menezes L.H.S., Soares G.A., dos Santos Reis N., Tavares I.M.C., Franco M., de Oliveira J.R. Production and biochemical characterization of halotolerant β-glucosidase by Penicillium roqueforti ATCC 10110 grown in forage palm under solid-state fermentation. Biomass Convers. Biorefin. 2020:1–12. doi: 10.1007/s13399-020-00930-8. [DOI] [Google Scholar]
- 36.Monteiro L.M.O., Vici A.C., Pinheiro M.P., Heinen P.R., de Oliveira A.H.C., Ward R.J., Prade R.A., Buckeridge M.S., Polizeli M.d.L.T.d.M. A highly glucose tolerant ß-glucosidase from Malbranchea pulchella (MpBg3) enables cellulose saccharification. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-63972-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fusco F.A., Fiorentino G., Pedone E., Contursi P., Bartolucci S., Limauro D. Biochemical characterization of a novel thermostable β-glucosidase from Dictyoglomus turgidum. Int. J. Biol. Macromol. 2018;113:783–791. doi: 10.1016/j.ijbiomac.2018.03.018. [DOI] [PubMed] [Google Scholar]
- 38.Xie J., Xu H., Jiang J., Zhang N., Yang J., Zhao J., Wei M. Characterization of a novel thermostable glucose-tolerant GH1 β-glucosidase from the hyperthermophile Ignisphaera aggregans and its application in the efficient production of baohuoside I from icariin and total epimedium flavonoids. Bioorgan. Chem. 2020;104 doi: 10.1016/j.bioorg.2020.104296. [DOI] [PubMed] [Google Scholar]
- 39.Ikai A. Thermostability and aliphatic index of globular proteins. J. Biochem. 1980;88:1895–1898. doi: 10.1093/oxfordjournals.jbchem.a133168. [DOI] [PubMed] [Google Scholar]
- 40.Kaur A., Pati P.K., Pati A.M., Nagpal A.K. Physico-chemical characterization and topological analysis of pathogenesis-related proteins from Arabidopsis thaliana and Oryza sativa using in-silico approaches. PLoS One. 2020;15 doi: 10.1371/journal.pone.0239836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 42.Graebin N.G., Schöffer J.d.N., Andrades D.D., Hertz P.F., Ayub M.A., Rodrigues R.C. Immobilization of glycoside hydrolase families GH1, GH13, and GH70: state of the art and perspectives. Mol. 2016;21:1074. doi: 10.3390/molecules21081074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H.Y., Chen Q., Zhang Y.F., Chen D.D., Yi X.N., Chen D.S., Cheng X.P., Li M., Wang H.Y., Chen K.Q. Improving the catalytic activity of β-glucosidase from Coniophora puteana via semi-rational design for efficient biomass cellulose degradation. Enzym. Microb. Technol. 2023;164 doi: 10.1016/j.enzmictec.2022.110188. [DOI] [PubMed] [Google Scholar]
- 44.Khairudin N.B.A., Mazlan N.S.F. Molecular docking study of beta-glucosidase with cellobiose, cellotetraose and cellotetriose. Bioinformation. 2013;9:813. doi: 10.6026/97320630009813. 10.6026%2F97320630009813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramachandran P., Tiwari M.K., Singh R.K., Haw J.-R., Jeya M., Lee J.-K. Cloning and characterization of a putative β-glucosidase (NfBGL595) from Neosartorya fischeri. Process Biochem. 2012;47:99–105. doi: 10.1016/j.procbio.2011.10.015. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.