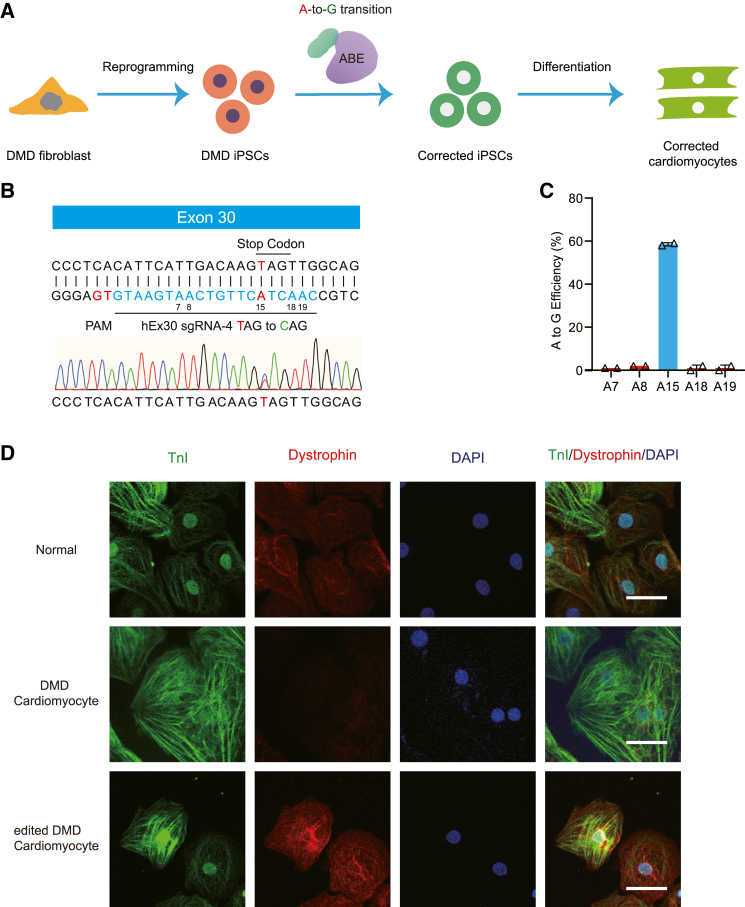
Figure 2.
Adenine base editing rescues dystrophin expression in human DMD cardiomyocytes
(A) Illustration of the application of adenine base editing in human DMD iPSC model. Human DMD iPSCs from patient fibroblasts were edited by the SpG-ABE components and subsequently differentiated into cardiomyocytes for downstream analyses. (B) Schematic for the binding position of hEx30 sgRNA-4 in the exon 30 mutant. The PAM and sgRNA are shown in red and blue respectively. The adenines in the editing window are numbered from the PAM. Genomic sequencing chromatogram of the exon 30 mutant in human iPSCs is present at the bottom. (C) The percentages of genomic editing events in human DMD iPSCs with ABE and hEx30 sgRNA-4 treatment (n = 3). On-target adenine (A15) is shown in blue. (D) Immunofluorescence analysis of dystrophin and cTnI levels in the normal, DMD, and corrected cardiomyocytes. Dystrophin and cTnI are shown in red and green, respectively. Nuclei are marked by DAPI stain in blue. Scale bar, 50 μm. Quantitative data are presented as mean ± SEM.