Figure 3.
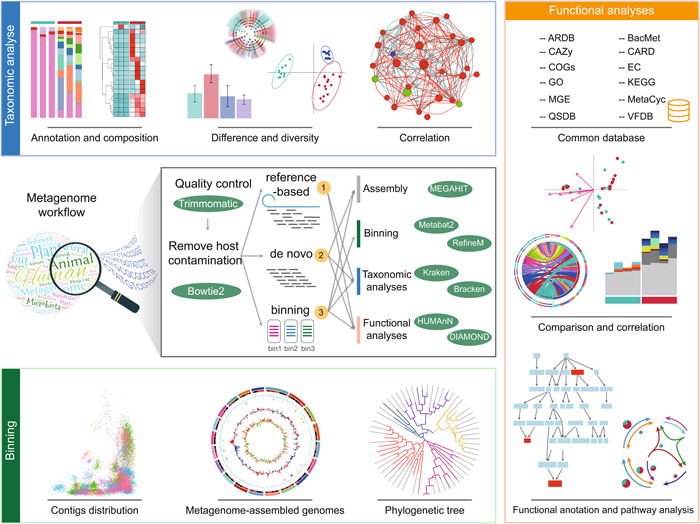
Metagenomic data analyses using Wekemo Bioincloud platform. The clean data could be analyzed with three different workflows, the reference‐based, the de novo, and the binning. The main software and visualization during assembly, binning, taxonomic analyses, and functional analyses were presented, making it suitable for various purposes or scenarios. The functional analyses include 12 diversity databases. ARDB, Antibiotic Resistance Genes Database; BacMet, Antibacterial Biocide and Metal Resistance Genes; CARD, Comprehensive Antibiotic Resistance Database; CAZy, Carbohydrate‐Active EnZymes; COGs, Clusters of Orthologous Groups of proteins; EC, Enzyme Commission; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes; MetaCyc, Metabolic Pathway; MGE, Mobile Genetic Element; QS, Quorum Sensing; VFDB, Virulence Factors Database.
