FIGURE 1.
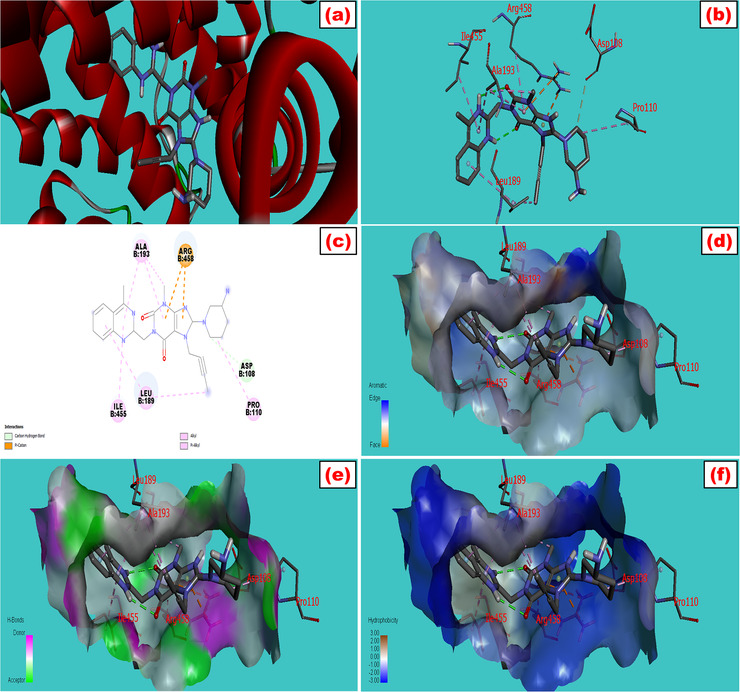
Molecular docking model of bovine serum albumin with (a) linagliptin which showed the best fitted geometric orientation of the ligand with protein. (b and c) 3D and 2D docking models of linagliptin with BSA, respectively. (d–f) The aromatic, hydrogen bond, and hydrophobic interactions across multiple side pockets of the protein, respectively. The various types of bonding were shown during interactions with their bond lengths. The SDF 3D conformer of the drug and PDB format of BSA (PDB: 3v03) were downloaded from PubChem and protein data bank, respectively. The ligand was docked with the protein by utilizing a set of extensive software like PyRx, PyMol 2.3, and Discovery Studio 4.5 to study the best‐fitted molecular orientation of LG with BSA. AutoDock Tools (ADT) and Swiss PDB viewer 4.1 were used to convert pdb into pdbqt format and to minimize the energy
