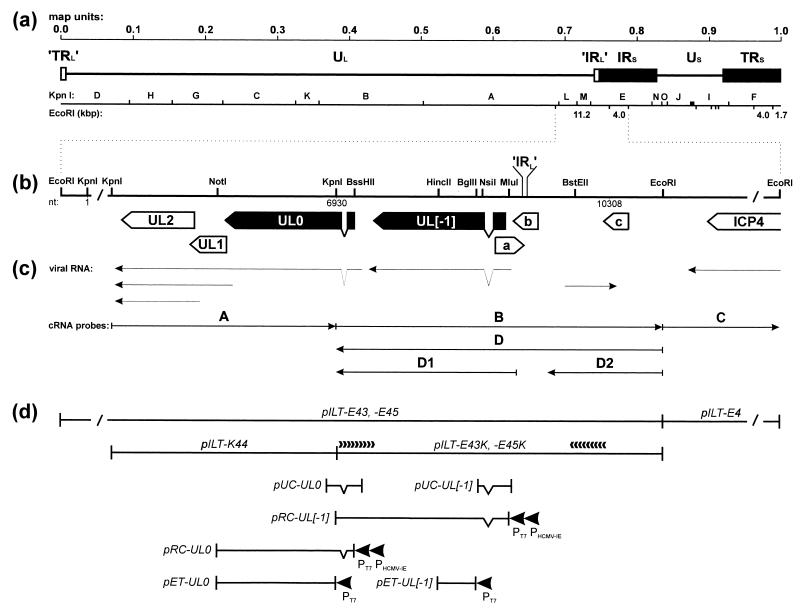
FIG. 1.
Genome structure of ILTV. (a) Diagram of the type D genome of ILTV with KpnI restriction fragment map. Locations and sizes of relevant EcoRI restriction fragments are indicated. The US region is flanked by extended inverted repeat sequences (IRS and TRS). Very short (17-bp) inverted repetitions (’IRL’ and ’TRL’) were also found at both ends of the UL region. (b) Enlarged map of the investigated region with relevant restriction sites. Close to either end, the DNA fragments are not plotted to scale (/). Location of the ’IRL’ sequence separating the UL and IRS regions is indicated. Nucleotide numbers refer to the DNA sequence deposited in GenBank (accession no. X97256). Locations and orientations of ORFs are depicted by pointed rectangles, and splice sites are indicated by triangles. Conserved genes are named according to their homologs in HSV-1, and the analyzed ILTV-specific genes are highlighted in black. (c) Arrows indicate identified viral transcripts and the in vitro-transcribed cRNA probes which were used for hybridization as shown in Fig. 2. (d) Plasmid maps. pILT-E43, -E45, -E4, and -K44 contain EcoRI or KpnI fragments of ILTV DNA. Plasmids pILT-E43K and -E45K were used for nested deletion subcloning and directed DNA sequencing as indicated by arrowheads. pUC-UL0 and pUC-UL[−1] are cDNA clones encompassing the 5′ termini of either mRNA. They were used for reconstitution of the processed UL0 and UL[−1] ORFs under control of the bacteriophage T7 (PT7) and HCMV immediate-early (PHCMV-IE) promoters in pRC-UL0 and pRC-UL[−1]. For immunization, bacterial fusion proteins were expressed from pET-UL0 and pET-UL[−1] under the control of the T7 promoter (PT7).