Figure 1 |. Precision genome editing in mammalian cells.
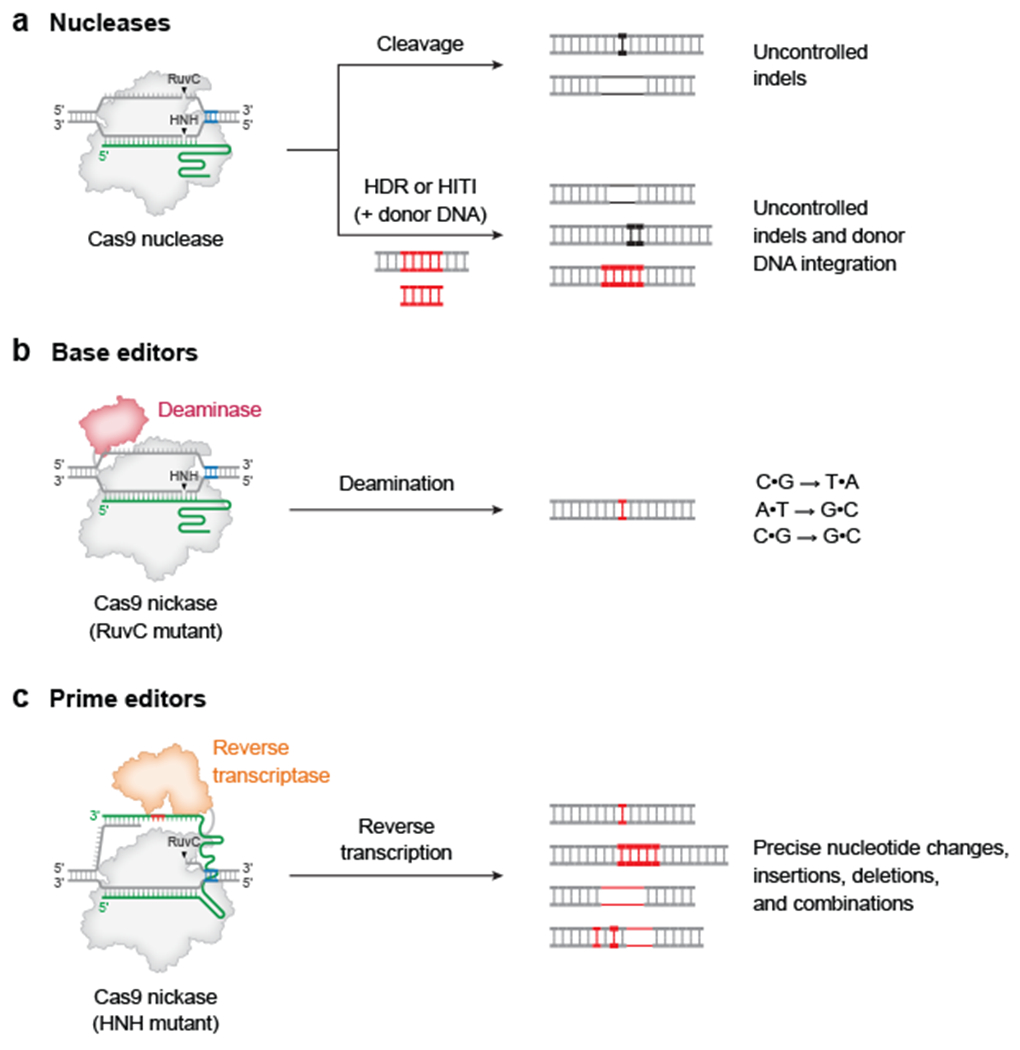
a | Cas nucleases can induce target DNA disrupt via formation of small insertions or deletions (indels), or DNA integration typically accompanied by substantial frequencies of undesired indel byproducts. b | Base editing can mediate C•G-to-T•A, C•G-to-G•C, and A•T-to-G•C conversions with few indel outcomes. Base editors canonically use a Cas9 nickase that only cuts the complementary strand. c | Prime editing can program any type of precise nucleotide substitutions, as well as insertions or deletions of up to hundreds of bases. Prime editors canonically use a Cas9 nickase that only cuts the non-complementary strand. Red DNA represents precisely edited sequence, and black DNA represents undesired outcomes. Blue DNA bases show the position of the protospacer-adjacent motif (PAM) required for Cas9 targeting. RuvC and HNH represent nuclease domains of Cas9. HDR, homology-directed repair; HITI, homology-independent targeted integration
