Figure 4 |. Prime editing variants.
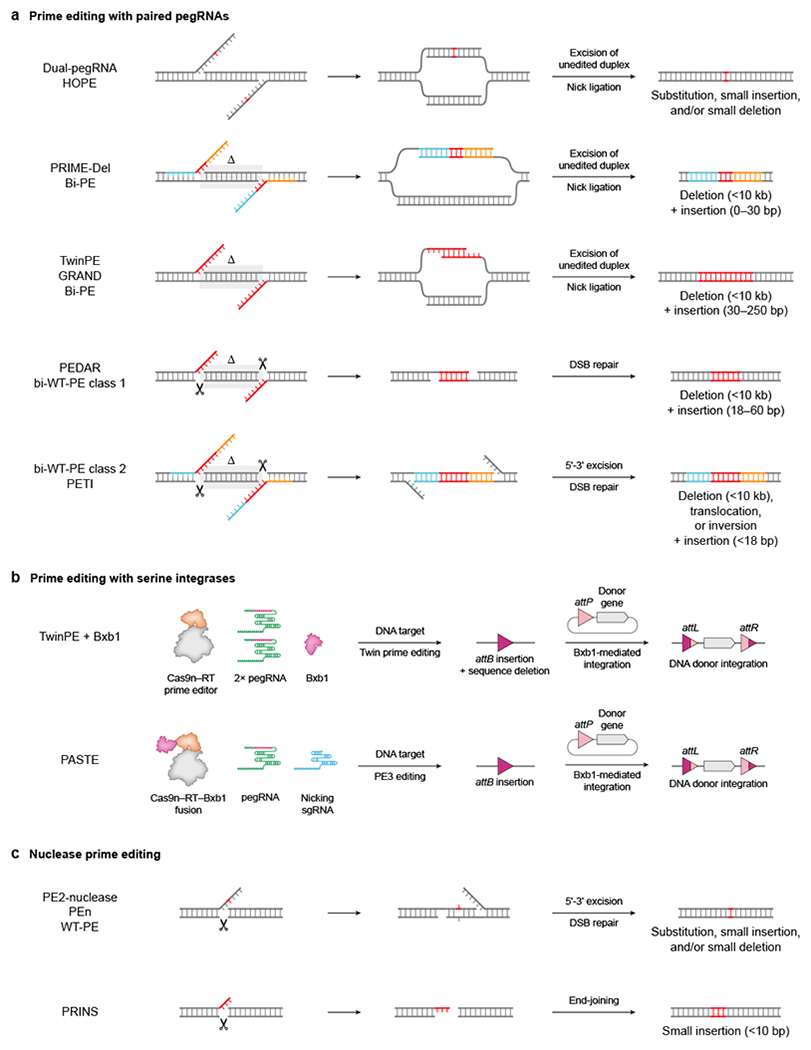
a | Prime editing with paired prime editing guide RNAs (pegRNAs) that template two 3ʹ DNA flaps containing the edit55–57,99,101–105. DNA intermediates formed after prime editor (PE)-mediated nicking and flap extension at the pegRNA target sites are shown on the left. Annealing of the complementary flaps, excision of the original genomic duplex, and ligation of the resulting nicks can efficiently mediate small edits or large insertions and deletions. b | Prime editing with serine integrases enables the targeted insertion of gene-sized (>1 kilobase) DNA segments. Twin prime editing or PASTE first installs an attB site into the genome using prime editing, then uses Bxb1 recombinase to mediate integration of donor DNA into the site56,58. c | Prime editors containing Cas9 nuclease create double-strand breaks (DSBs) with an edited 3ʹ overhang that can be incorporated into the genome but also result in frequent formation of small insertion or deletion (indel) byproducts104,106,107. Red DNA bases represent edited DNA with heterologous sequence. Blue and orange DNA bases represent homologous sequence. ∆ specifies DNA bases deleted by the edit. Scissors denote sites of Cas9-induced DSBs.
