Figure 1.
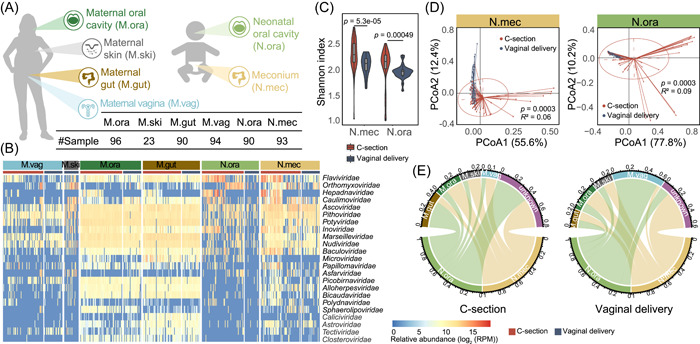
Sample collection and virome comparison between C‐section and vaginally delivered neonates and their mothers. (A) Samples were collected from six body sites, including oral (M.ora), intestinal (M.gut), vaginal (M.vag) and skin (M.ski) of mothers, and oral (N.ora, oral contents collected within seconds after birth) and intestinal (N.mec, meconium, i.e., the first excretion after birth) of newborns. The number of each sample type is listed in the table. (B) Relative abundance of viruses in each body site is profiled at the family level. The red and blue bars represent samples of subjects delivered with C‐section and vaginal mode, respectively. The virus families with the largest intergroup variance (the intergroup variance of abundance is greater than the overall median variance) are shown. Since no skin samples of C‐section mothers were collected, only the M.ski of vaginal delivery is used here. (C) Alpha diversities of viruses in N.mec (left) and N.ora (right) were calculated at the family level and are colored by delivery modes. Violins show median (black horizontal line), quartiles (edge of boxes), and kernel density estimation (violin) for each distribution. (D) Principal co‐ordinates analysis (PCoA) of virome profiles in N.mec (left) and N.ora (right) was performed at the family level. (E) Viral source tracking from mothers to neonates delivered by C‐section (left) and vagina (right), respectively. The proportion of shared viruses between generations was measured at the species level.
