Figure 1.
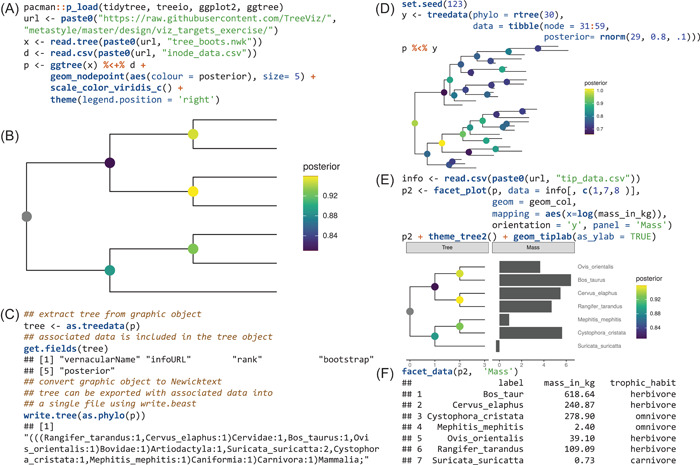
Examples of using ggtree object to store phylogenetic tree, associated data, and visualization directive. A tree in Newick format was read into R as x and an associated data, d, that stores posterior of internal nodes were used to visualize circle points that were colored by the posterior values. The output graphic object, that is, the ggtree object, was stored in p (A). The ggtree object, p, can be rendered as a picture by print(p) (B). The ggtree object contains the information of the phylogenetic tree and associated data. It can be converted back to a treedata object with the tree and all the associated data, including posterior, bootstrap, and other values in d that were attached to the graphic object, p. The tree can be exported to Newick text (without associated data) and BEAST Nexus file (with associated data) (C). The ggtree object contains visualization directives that can be reused to visualize a new tree object by applying its visualization style (D). Associated data can be visualized with the tree side by side using facet_plot (E). The data used in facet_plot can also be extracted from the graphic object using facet_data function (F).
