Figure 2.
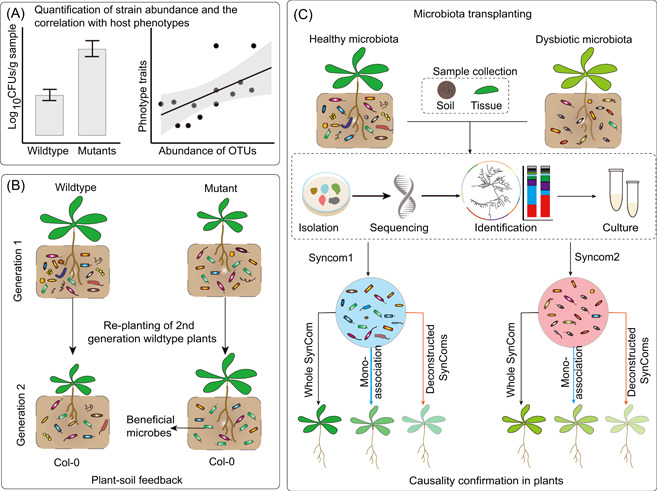
Approaches to link microbiota changes and casual phenotypes in plants. (A) For mutants affecting Pseudomonas or other microbes that are easy to isolate and quantify, we can quantify the CFUs directly in different groups. If the abundance of strains of interest is correlated with the plant traits (fresh weight or other stress‐related measurements), this can provide primary clues about the linkage between microbial changes and plant phenotypes. For most bacterial strains that cannot be isolated and characterized easily, we can analyze the correlation based on the relative abundance from microbiome sequencing data or PCR quantification of species‐specific marker genes. (B) A plant–soil feedback system can be used to confirm whether a changed microbiota would lead to specific phenotypes (in the diagram: growth promotion) in plants. Green bacteria indicate beneficial and growth‐promoting bacteria, which were enriched by the first‐generation plants. (C) The SynCom‐based microbiota transplantation assay could be used to confirm the causal phenotype of microbiota changes. The diagram shows that a dysbiosis causes pale and sick phenotypes in plants. To confirm the causal link, we can isolate strains from both the healthy and dysbiotic communities and perform whole‐SynCom transplantation to determine whether transplantation of the dysbiotic microbiota phenocopies the original plants with a dysbiotic microbiota. The SynCom‐based approach also provides powerful flexibility for performing either single‐strain (mono‐association) screening or combining strain modules of interest. CFU, colony‐forming units; PCR, polymerase chain reaction
