Figure 1.
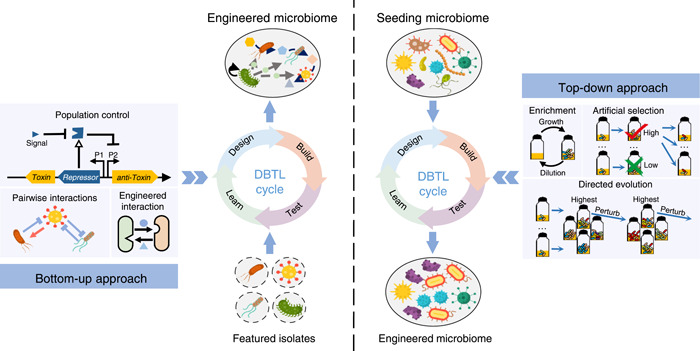
Two approaches to microbiome engineering. Left panel: the bottom‐up approach of microbiome engineering starts with several isolates, whose physiologic features have been well characterized. Coculture assays are performed to identify pairwise interactions among these isolates. Genetic engineering could be performed to design or modify the interactions. Systems based on cell‐to‐cell communications are used to directly control the behavior of specific populations. Right panel: the top‐down approach of microbiome engineering starts with a seeding microbiome containing uncultivated microorganisms. Then, microbiome engineering is managed to drive the seeding microbiome to self‐assemble into a stable system with highly optimized function. Three methods, including enrichment, artificial selection, and directed evolution, are commonly applied to perform top‐down engineering. No matter which approaches are used, the workflows should follow the “design‐build‐test‐learn” (DBTL) cycle proposed by Lawson et al. 13 .
