Figure 4.
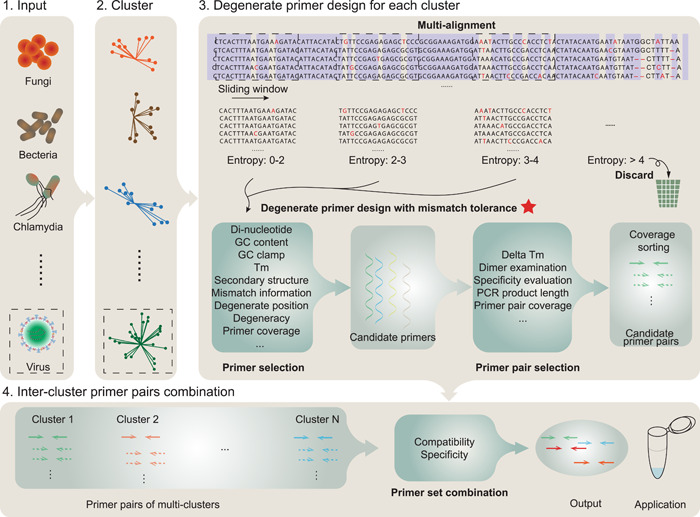
Schematic diagram depicting the one‐step targeted next‐generation sequencing (tNGS) primer set design method developed by using multiPrime. The red star indicates the step where the method is described in more detail. MultiPrime comprises four primary stages: (1) Input: In this initial phase, the collection of target sequences is required. (2) Cluster: Redundant sequences are eliminated and clusters are established based on sequence identity. (3) Degenerate Primer Design for Each Cluster: Utilizing MUSCLE or MAFFT, a multialignment procedure is conducted, followed by the design of candidate primers using the nearest‐neighbor model. (4) Intercluster Primer Pair Combination: Primer pairs are selected considering factors such as PCR product length, melting temperature, dimer formation assessment, coverage despite errors, and other pertinent criteria. Subsequently, a greedy algorithm is employed to merge primer pairs into an optimal minimal primer set, guided by dimer formation analysis.
