Figure 1.
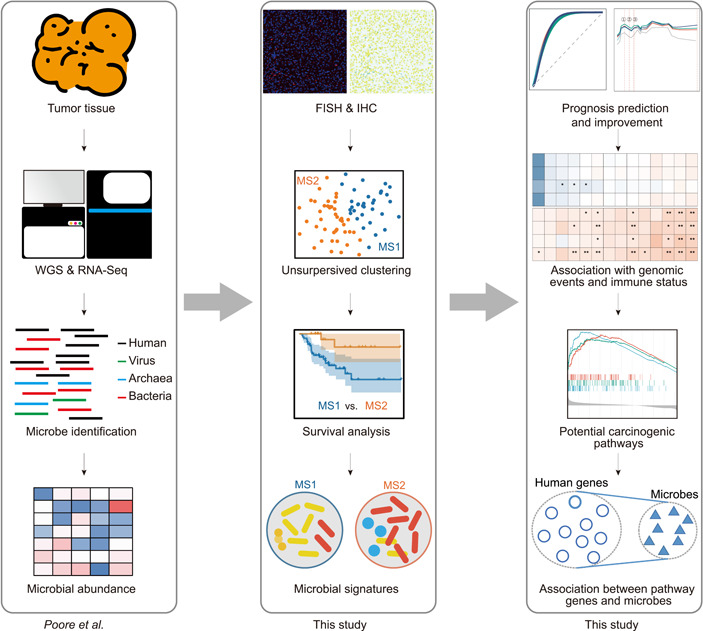
Overview of the analysis pipeline. The Cancer Genome Atlas primary tumors were subject to whole‐genome sequencing and RNA‐Seq, in which microbial abundance (comprising virus, archaea, and bacteria) was identified and normalized by Poore et al. (left panel). To confirm the presence of intratumoral bacteria, 37 adrenocortical carcinoma tissue microarray chips from the in‐house cohort were stained for fluorescence in situ hybridization and immunohistochemistry examination. Unsupervised clustering was performed on microbiome data to explore natural clusters of patients with distinct microbiomes, thereby influencing prognoses. The microbial signatures associated with prognosis were further determined (middle panel). Defined microbial signatures were tested to improve prognosis prediction. The intratumoral microbiome was found to be associated with immune status, genetic events, and molecular pathways (right panel).
