Fig. 4.
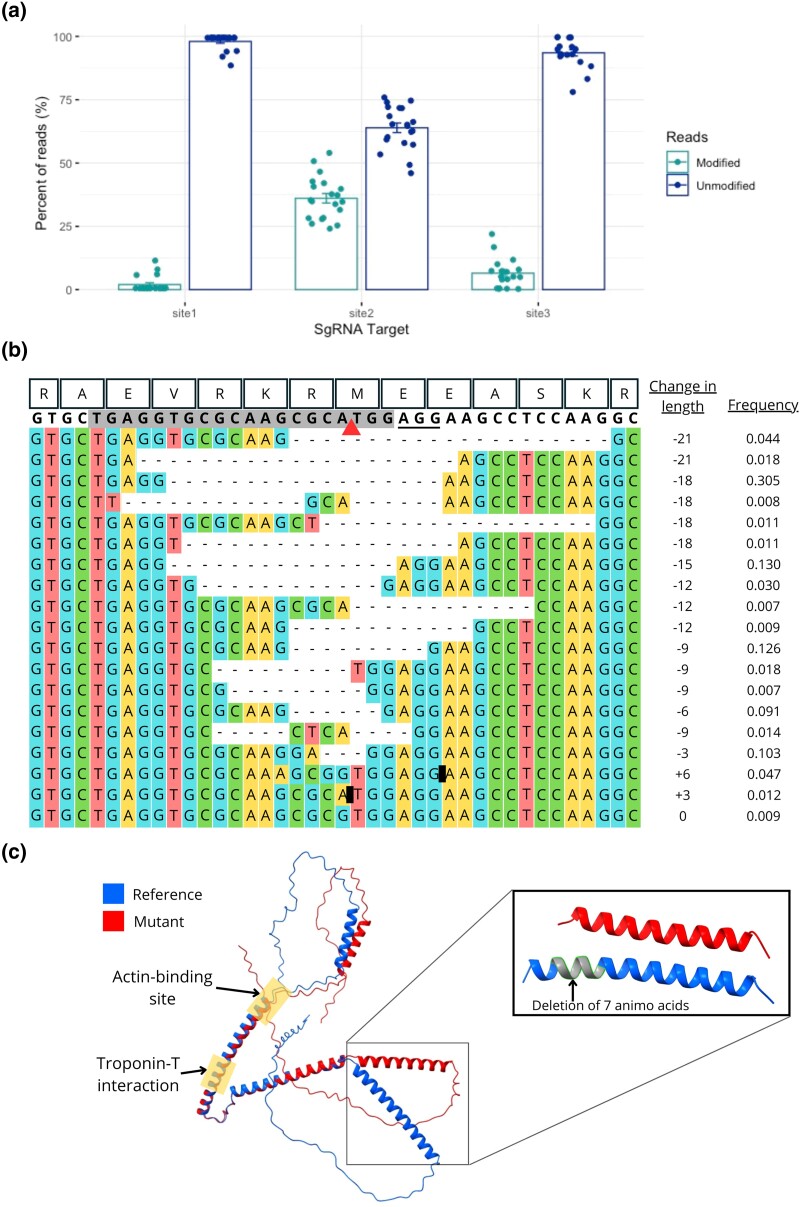
Recessive lethal wupA mutations revealed in females. a) The 3 sites within wupA that are targeted by sgRNAs (see Fig. 1c) vary in the number of CRISPR mutations uncovered by sequencing 200 daughters of an X-poisoning cross. Here, a CRISPResso2 output shows the total number of modified and unmodified reads from Illumina sequencing at each target site. b) A nucleotide alignment of the modified reads of target site 2 showing their frequencies. Amino acid sequences are shown at the top, the sgRNA sequence is highlighted in gray, the PAM site is underlined, the triangle is the cut site, and the black blocks represent insertions (shown more to detail in Supplementary Fig. 3). c) An overlay of Isoform G of troponin I without any mutations (blue) and with the largest deletion found in our sequencing results (a 21 bp deletion shown in red). Protein folding predicted with AlphaFold. The deletion location is highlighted in gray. No mutations in target site 2 will affect the acting-binding and troponin-interacting areas.