Figure 1.
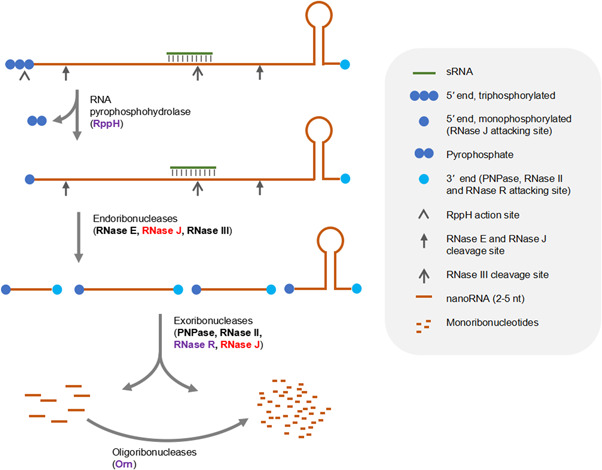
The principal pathway of messenger RNA (mRNA) degradation in bacteria. The major steps include the following: RNA phosphohydrolase converts the 5ʹ end of the primary transcripts from triphosphate to monophosphate; endoribonucleases internally cleave the transcripts into intermediate fragments; exoribonucleases degrade the intermediates into monoribonucleotides and generate the end products of 2‐ to 5‐nt oligoribonucleotides (nanoRNAs); and oligoribonucleases hydrolyze the oligoribonucleotides into monoribonucleotides, finishing the degradation process. Note that many transcripts in the triphosphorylated form can also be substrates of endoribonucleases and some transcripts may also be substrates of exoribonucleases before endoribonucleolytic cleavage. The major Escherichia coli and cyanobacterial enzymes involved in the degradation process are shown. The enzymes present in both E. coli and cyanobacteria (RNase E, RNase III, PNPase, and RNase II) are in black, those currently discovered in E. coli only (RppH, RNase R, and Orn) are in purple, and the one present in cyanobacteria but not in E. coli (RNase J) is in red. Note that RNase J acts as both an endoribonuclease and a 5ʹ−3ʹ exoribonuclease.
