Figure 3.
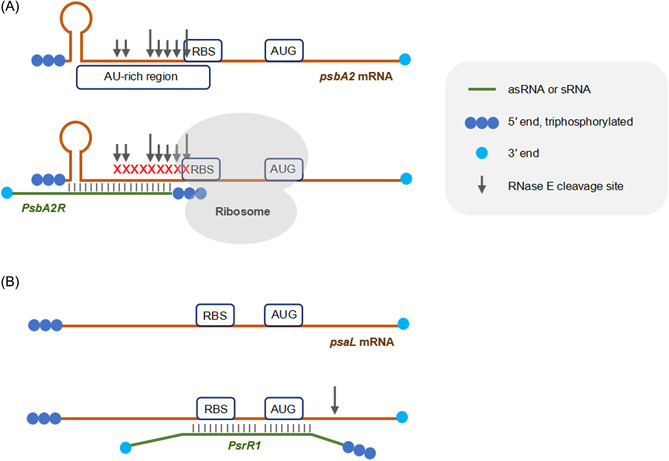
RNase E cleavage of messenger RNAs (mRNAs) encoding photosynthetic proteins in Synechocystis PCC 6803. (A) Upper panel: The psbA2 mRNA has a 49‐nt 5ʹ UTR that, in the dark, is sensitive to RNase E cleavage due to multiple RNase E cleavage sites within an AU‐rich sequence that extends into the likely ribosome binding site (RBS) 43 , 50 , 57 . Hence, rapidly interacting ribosomes under conditions such as high light, when this mRNA is strongly translated, can protect against degradation (lower panel). In addition, the antisense RNA (asRNA) PsbA2R is coregulated with the mRNA from a transcriptional start site leading to an overlap with the first 19 nucleotides of the psbA2 mRNA, hence protecting these sites against RNase E cleavage 58 , as indicated by the red crosses. A short stem‐loop near the mRNA 5ʹ end might be relevant for recognition. Note that in vivo additional ribonucleases are likely involved 50 . Note that the gene psbA3, which is almost identical to psbA2, is regulated in the same way 57 , 58 . (B) Upper panel: The psaL gene is preceded by a 55‐nt 5ʹ UTR that is not targeted by RNase E under most growth conditions. However, under high light, transcription of the gene for the sRNA PsrR1, which is located elsewhere in the genome, is stimulated. Except for two mismatches, PsrR1 interacts over 22 consecutive nucleotides with the psaL mRNA (lower panel). This interaction overlaps the 3ʹ end of the 5ʹ UTR, including the ribosome binding site, the start codon, and one additional nucleotide of the second codon. This interaction leads to conditional recruitment of RNase E, which then cleaves a single‐bond seven nucleotides into the coding sequence, effectively decapitating the psaL mRNA 59 .
