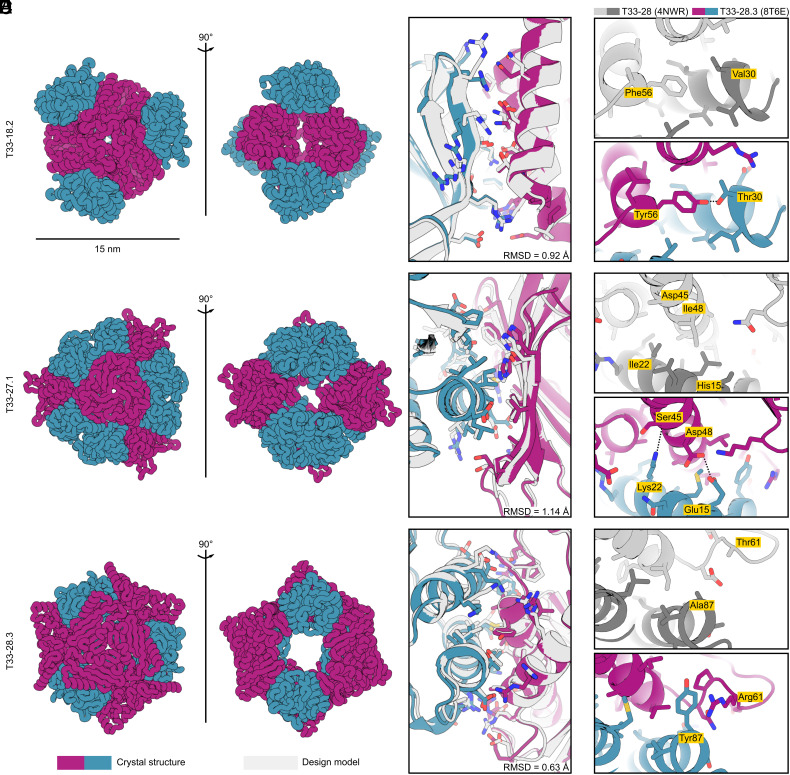
Fig. 5.
Crystal structures of ProteinMPNN-designed tetrahedral nanoparticles. (A) Ribbon displays of crystal structures of three tetrahedral nanoparticles: T33-18.2 (PDB ID: 8T6C), T33-27.1 (PDB ID: 8T6N), and T33-28.3 (PDB ID: 8T6E). (B) Atomic interactions at the designed interfaces. Side chains of residues with atoms within 5.5 Å of the opposing subunit are shown as sticks. Light gray color indicates the ProteinMPNN design model and purple-blue colors indicate the crystal structure of components A and B, respectively. In all three instances, the crystal structures matched the design models with high accuracy, with backbone RMSD over the ASU of: 0.92 Å (T33-18.2), 1.14 Å (T33-27.1), and 0.63 Å (T33-28.3). (C–E) Comparison of selected interface interactions in the crystal structures for T33-28 (PDB ID: 4NWR; ref. 20) and T33-28.3 (PDB ID: 8T6E). (C) In T33-28.3A the hydrophobic Phe56 is replaced with a Tyr with its hydroxyl group hydrogen bonding with Thr33 on T33-28.3B. (D) T33-28.3 features two interface stabilizing polar interactions at the core. The Lys22 amino group forms a hydrogen bond with the backbone carbonyl of Ser45, and Asp48 forms hydrogen bonds with Glu15. In contrast, the core positions for T33-28 (Ile22 and Ile48) featured exclusively hydrophobic packing. (E) In a poorly packed region of T33-28, T33-28.3 features a cation-pi interaction between Tyr87 and Arg61. Note: in the crystal structure of T33-28, only the Cβ atom of the His15, Asp45, and Thr61 side chains were resolved.