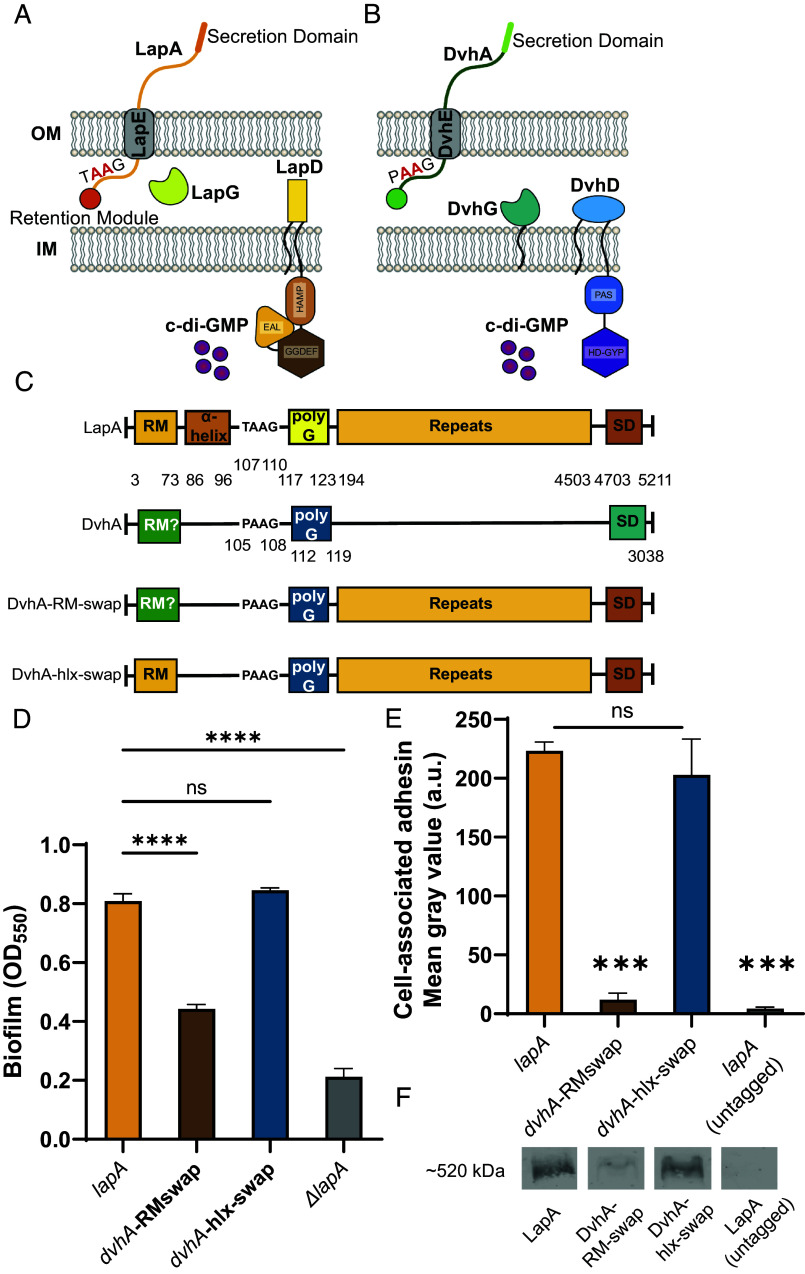
Fig. 1.
Simplified model of two-step Type 1 secretion system (T1SS). (A) T1SS in P. fluorescens Pf0-1. The large adhesin LapA is localized to the outer membrane (OM) via a retention module, allowing the bacterium to form a biofilm. The localization of LapA to the OM is regulated by a periplasmic protease LapG and a cyclic di-GMP (cdG) effector protein LapD. At high intracellular cdG levels, the catalytically inactive EAL domain of LapD binds cdG leading to a conformational change whereby the LapD periplasmic domain sequesters LapG, thereby retaining LapA on the OM. When c-di-GMP levels are low, LapD is in its autoinhibited conformation (as shown here), which releases LapG to cleave LapA at a di-alanine motif (shown in red), resulting in the release of LapA to the extracellular environment and loss of biofilm formation. (B) Proposed model of T1SS in D. vulgaris Hildenborough. DvhA is a LapA-like adhesin localized to the OM. DvhG is a LapG-like protease that can cleave DvhA at the di-alanine site (shown in red), however, unlike the periplasmic-localized LapG, DvhG is hypothesized to be inner membrane (IM) bound. DvhD is a structurally different but functionally analogous LapD-like protein containing a catalytically inactive HD-GYP domain that can bind c-di-GMP and thus regulate DvhA localization and biofilm formation by D. vulgaris Hildenborough. (C) Simplified protein architecture representing different domains and the di-alanine site of Pf0-1 adhesin LapA, DvH adhesin DvhA and fusion proteins DvhA-RM-swap and DvhA-hlx-swap (RM-retention module; hlx-helix; polyG-polyglycine linker; SD – C-terminal secretion domain). Protein domains are not to scale. (D) Biofilm formed in K10T medium at 24 h for strains expressing the adhesins DvhA-RM-swap and DvhA-hlx-swap compared to the native adhesin LapA in Pf0-1 and the ΔlapA mutant, all in the ΔlapGΔlapD background strain. (E) Quantification of cell surface levels of HA-tagged adhesins at 24 h in K10T medium for the indicated strains. (F) Western blots indicating total cellular levels of the different adhesins. Statistical analysis was performed using one-way ANOVA corrected for multiple comparisons (ns, P > 0.05; *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001; ****P ≤ 0.0001; ****P < 0.0001).