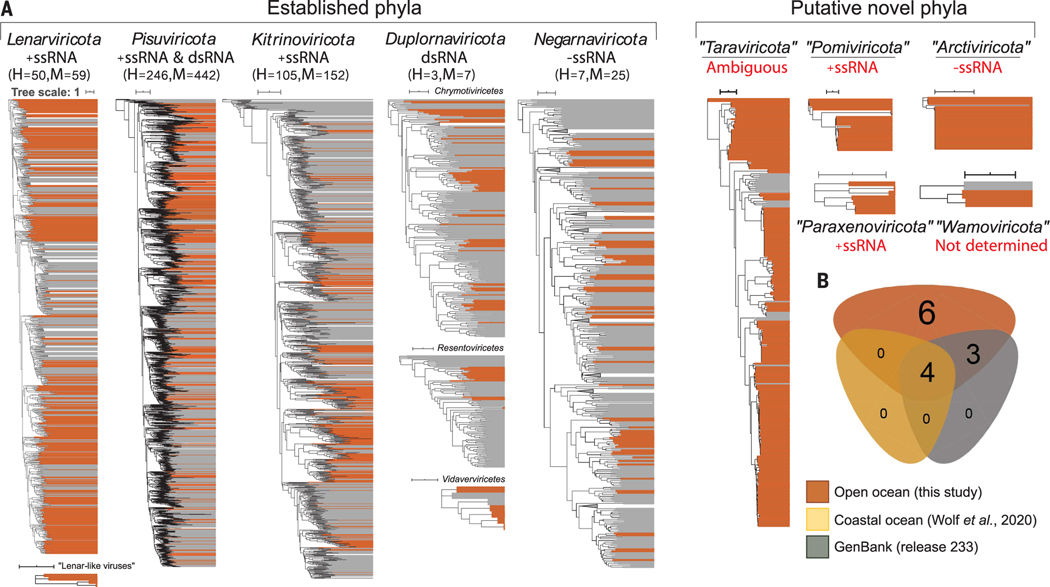
Fig. 2. Phylum- and class-rank RdRp-based phylogenetic analyses showing the taxonomic diversity of Global Ocean orthornavirans.
(A) Thirteen maximum-likelihood phylogenetic trees encompassing the 19 megaclusters that emerged from network analyses of near-complete RdRp sequences (details in Fig. 1). Brown color indicates virus sequences discovered in this study, whereas gray indicates previously known reference sequences. The scale bar indicates one amino acid substitution per site. Classes were merged into a unified phylum-ranked tree only if the results from both phylogeny and network-guided clustering analysis were in agreement (materials and methods). Sequences were preclustered at 50% identity, and clades supported by 100% bootstrap values were collapsed. Genome strandedness (red text) for the new phyla was inferred in this study (as described in fig. S8 and materials and methods). A conservative estimate of the number of new complete or high-quality (H) and medium-quality (M) genomes retrieved in this study is indicated with parentheses. Underlined new phyla are supported by long- and short-read assemblies, whereas the remainder were supported by multiple independent assemblies from short-read assemblies (domain motifs are available in table S10). (B) Euler diagram of the shared, well-resolved phylum-or class-rank clusters of the near-complete RdRp domains across all available data from GenBank, a prior coastal ocean survey, and this study. Established megataxa represented in all datasets are Lenarviricota, Pisuviricota, Kitrinoviricota, and Duplornaviricota; Chrymotiviricetes. Established megataxa represented in our dataset and GenBank are Duplornaviricota; Vidaverviricetes, Duplornaviricota; Resentoviricetes, and Negarnaviricota. Unestablished megataxa inferred in this study are “Taraviricota,”“Pomiviricota,” “Paraxenoviricota,”“Arctiviricota,”“Wamoviricota,” and “lenar-like viruses.” In all analyses, RdRp domain clusters with permuted motifs (“permutotetra-like” and “birna-like” viruses) were excluded.