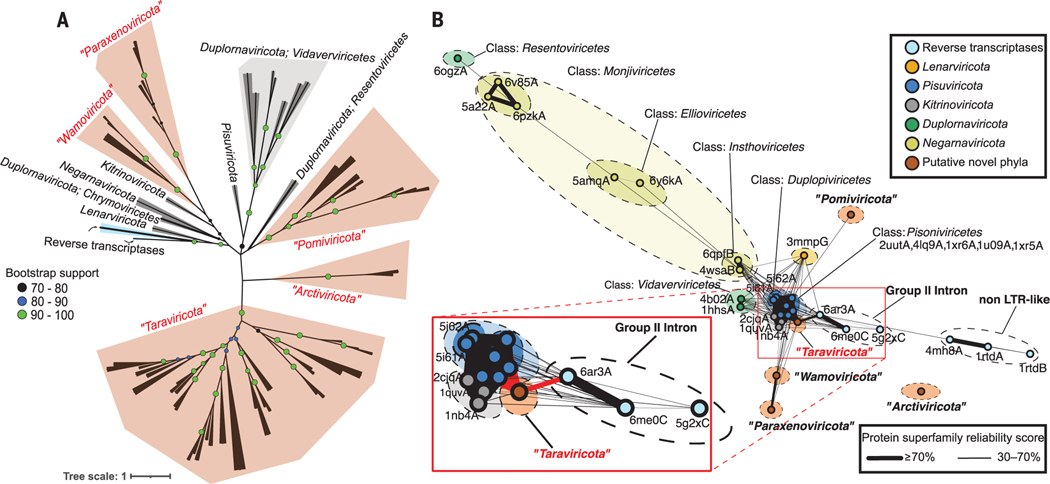
Fig. 3. Global RdRp-based phylogeny and network analyses inferring the early evolutionary history of orthornavirans.
(A) Maximum-likelihood phylogenetic tree of RdRp domain sequences with RT sequences (cyan). The gray branches and polygons represent established megataxa, whereas the brown polygons represent megataxa inferred here. Each branch represents either a consensus or an individual sequence from a megataxon (materials and methods). Nodes in each branch represent bootstrap support. The scale bar indicates one amino acid substitution per site. (B) Three-dimensional structure similarity network of predicted (brown) and experimentally resolved (other colors; labeled with accession numbers) RdRp and RT protein domain structures. Each node represents a different structure, and the edges represent the reliability scores, for each connected pair, that they belong to the same protein superfamily (materials and methods). (Inset) The probability of “taraviricot” RdRps belonging to the same superfamily as group II–intron RTs and pisuviricot RdRps is 75 and 98%, respectively. In all analyses, RdRp domain clusters with permuted motifs (“permutotetra-like” and “birna-like” viruses) were excluded. LTR, long terminal repeat.