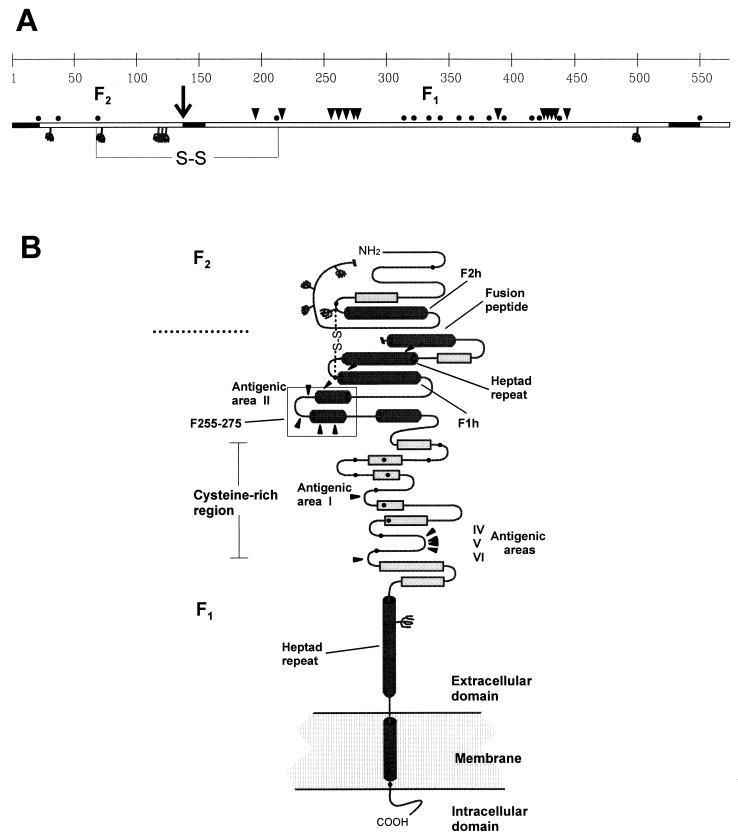
FIG. 4.
Prediction of secondary-structure motifs of the F protein. (A) Linear diagram of the F protein polypeptide of the Long strain (22), showing cysteine residues (•), potential N-glycosylation sites ( ), site of proteolytic processing (↓), amino acid changes in escape mutants reported here and in previous publications (▾) and hydrophobic regions (▪). S-S, disulfide bridge. (B) Predicted secondary-structure elements of the F protein: α-helices (cylinders), β-sheets (rectangles), loops (turns). Other symbols are as explained for panel A. The segment between residues 255 and 275 (F255-275), predicted to fold in a helix-loop-helix structure, is boxed. F1h and F2h, amphipathic α-helices in the F1 and F2 chains, respectively. The diagram is not drawn to scale. The predictions were made with published sequences of pneumovirus F proteins and by using the following methods available on Worldwide Web servers: the PHD method (33, 34) (http://www.embl-heidelberg.de/predictprotein/predictprotei.html), the DSC method (18) (http://bonsai.lif.icnt.uk/bmm/dsc_form_align.html), and the PREDATOR method (11) (http://embl-heidelberg.de/predator/predator_info.html), which use multiple aligned protein sequences as input to estimate the probability of secondary-structure motifs and solvent accessibility; and the SOPMA method (13) (http://www.ibcp.fr/serv_pred.html), the nnPREDICT method (19) (http://www.cmpharm.ucsf.edu/~nomi/nnpredict.html), and the PSA method (38) (http://bmerc-www-bu.edu/psa/), which use single protein sequences as input. The final secondary-structure model was a consensus of the information obtained by the different methods. Details of the prediction studies are available from the authors upon request.
), site of proteolytic processing (↓), amino acid changes in escape mutants reported here and in previous publications (▾) and hydrophobic regions (▪). S-S, disulfide bridge. (B) Predicted secondary-structure elements of the F protein: α-helices (cylinders), β-sheets (rectangles), loops (turns). Other symbols are as explained for panel A. The segment between residues 255 and 275 (F255-275), predicted to fold in a helix-loop-helix structure, is boxed. F1h and F2h, amphipathic α-helices in the F1 and F2 chains, respectively. The diagram is not drawn to scale. The predictions were made with published sequences of pneumovirus F proteins and by using the following methods available on Worldwide Web servers: the PHD method (33, 34) (http://www.embl-heidelberg.de/predictprotein/predictprotei.html), the DSC method (18) (http://bonsai.lif.icnt.uk/bmm/dsc_form_align.html), and the PREDATOR method (11) (http://embl-heidelberg.de/predator/predator_info.html), which use multiple aligned protein sequences as input to estimate the probability of secondary-structure motifs and solvent accessibility; and the SOPMA method (13) (http://www.ibcp.fr/serv_pred.html), the nnPREDICT method (19) (http://www.cmpharm.ucsf.edu/~nomi/nnpredict.html), and the PSA method (38) (http://bmerc-www-bu.edu/psa/), which use single protein sequences as input. The final secondary-structure model was a consensus of the information obtained by the different methods. Details of the prediction studies are available from the authors upon request.