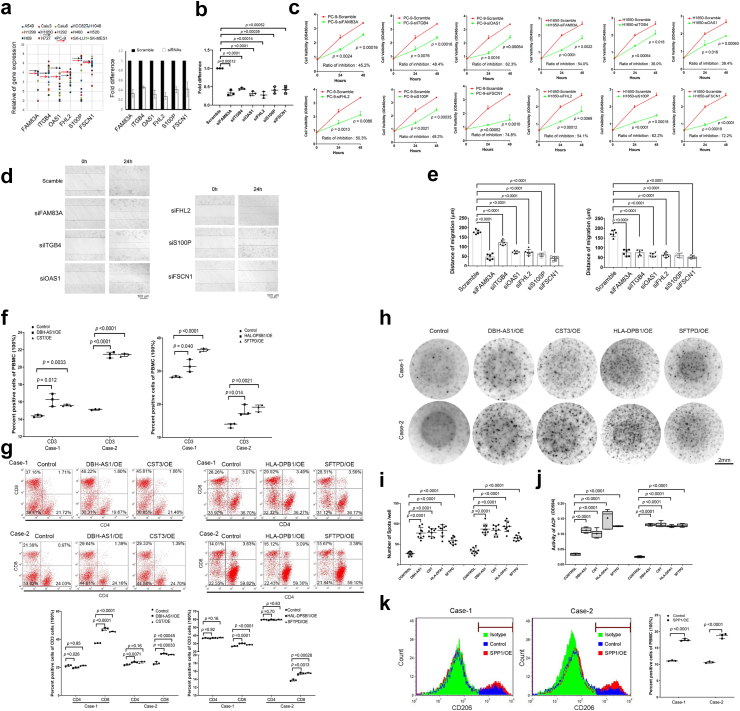
Fig. 3.
Results of In-vitro cell experiments. (a) The scatter plot shows the expression difference of multiple genes in multiple lung cancer cell lines. The black arrow represents H1650 and the red arrow represents PC9. (b) The fold change of the gene expression following multiple gene respectively knockdown in PC-9 cells was confirmed by RT-PCR. (c) The cell variability curve demonstrated a significant decrease in cell survival following genes knockdown in PC9 and H1650. (d) Wounding assay shows cells movement following genes knockdown after 24-h. Scale Bar = 100 μm. (e) The bar chart shows the distance of migration by the cells of genes knockdown in PC-9 and H1650. (f) The flow cytometry results revealed the presence of the percentage of CD3 cells in the peripheral blood mononuclear cells (PBMCs) of patients with overexpression of DBH-AS1, CST3, HLA-DPB1 and SFTPD. (g) The flow cytometry results the percentage of CD4 and CD8 in PBMCs of patients. (h) and (i) Elispots assay shows number of spots signature in DBH-AS1, CST3, HLA-DPB1 and SFTPD overexpressed cells of PBMCs. The horizontal lines denoted the mean quantity of spots per well. Scale Bar = 2 mm. (j) ELISA assay shows ACP activity in genes overexpressed cells of PBMCs. (k) The flow cytometry results revealed the presence of the percentage of CD206 cells in PBMCs with SPP1 overexpression. All experiments were conducted independently three times. The error bars refer to the standard deviation of the values. P values were determined by independent t-test. Two-tail paired t-test for b and c by mean with SD; One-way analysis of variance for e, i and j by mean with SD; Chi-square test for f, g and k by medium with 95% CI. Abbreviations: RT-PCR, reverse transcription-polymerase chain reaction; OE, overexpression model; siRNA, small interfering RNA; CI, confidence interval.