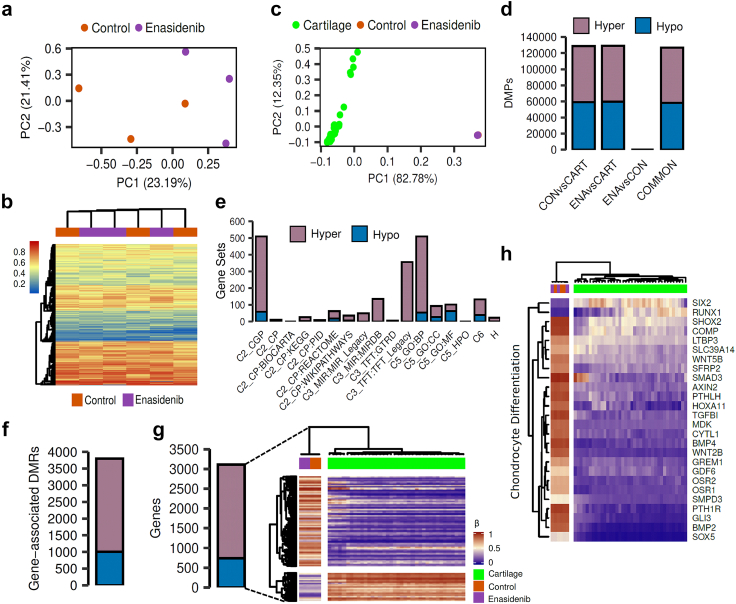
Fig. 4.
Effect of enasidenib on the methylome of IDH2-mutant chondrosarcoma cells. T-CDS-17#1 cells were treated in biological triplicates with DMSO (control; CON) or 20 μM enasidenib (ENA) for 48 h prior to be processed for DNA methylation analysis. (a) Scatter plot showing the PCA of CON and ENA samples according to the beta methylation values at the top 50,000 most variable CpG sites. (b) Heatmap diagram depicting the beta methylation values of CON and ENA samples at the top 5000 most variable CpG sites. No differentially methylated positions (FDR <0.05, |Δβ|>20%) between ENA and CON samples were found. (c–h) analysis of the methylation data of CON and ENA samples along with a public-available methylation datasets of healthy human cartilage samples (CART) (n = 39). (c) PCA of CON, ENA and cartilage samples according to the beta methylation values at the top 50,000 most variable CpG sites belonging to the HumanMethylation450K array. (d) Barplots depicting the number of hyper- and hypomethylated DMPs (FDR <0.05, |Δβ|>20%) found in CON vs CART, ENA vs CART and ENA vs CON comparisons. COMMON bar includes those hyper- and hypo- DMPs that are in both CON vs CART and ENA vs CART comparisons. (e) Barplots showing the number of significantly enriched pathways found using hyper- and hypomethylated common DMPs for different MSigDB gene sets. (f) Barplot depicting the number of hyper- and hypomethylated DMRs (FDR <0.05, |Δβ|>20%) found in gene regions using common DMPs. (g) On the left, a barplot showing the number of hyper- and hypomethylated genes. On the right, a heatmap plot showing the beta methylation values of the aforementioned differentially methylated genes. (h) Heatmap plot depicting the beta methylation values of those differentially methylated genes that belong to the GO BP chondrocyte differentiation pathway.