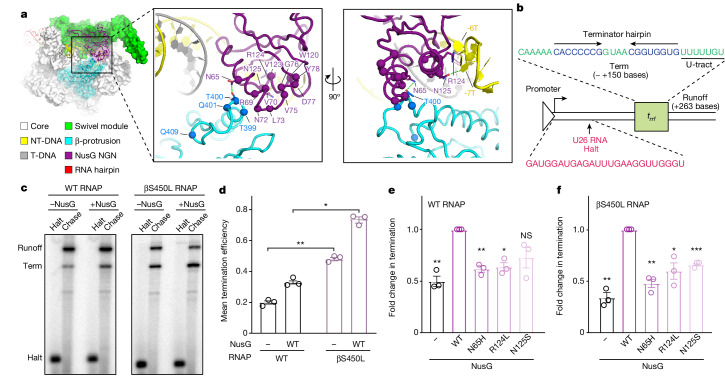
Fig. 4. Clinical strain mutations decrease the pro-pausing activity of NusG.
a, Cryo-EM structure of a NusG-bound paused elongation complex from Mtb. The boxed area shows the location of mutations observed in RifR clinical Mtb isolates. Interactions between NusG and the β-protrusion or non-template DNA are shown in green dashed lines13. b, Promoter template with the Mtb rrf termination sequence for in vitro transcription assays. The experimental schematic was followed for data shown in c–f. c, In vitro termination by Mtb RNAP on a promoter-initiated template with or without wild-type (WT) NusG. The U26, C-less halt site is labelled (Halt), as well as approximately +150 base termination site (Term) and +263 base runoff (Runoff). d, Termination efficiencies of wild-type and βS450L RNAP in the presence (two-sided adjusted P value = 0.039) or absence (two-sided adjusted P value = 0.002) of wild-type NusG. Data are mean ± s.d. from three experimental replicates. e, Fold change in termination (∆T) relative to wild-type RNAP plus wild-type NusG. Data are mean ± s.d. from three experimental replicates. ∆T for each NusG protein was calculated from kinetics of the elongation versus termination process as described by von Hippel and Yager62,63 based on the relationships between termination efficiency and the rates and activation energies of termination (Methods). Left to right, two-sided adjusted P value: 0.009, 0.006, 0.024 and 0.057. f, Fold change in termination as in e, relative to βS450L RNAP plus wild-type NusG. Left to right, two-sided adjusted P value: 0.009, 0.006, 0.038 and 8.7 × 10−4. (d–f) Independent two-sample t-test, with multiple hypothesis correction by Benjamini, Krieger and Yekutieli method. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001; NS, not significant.