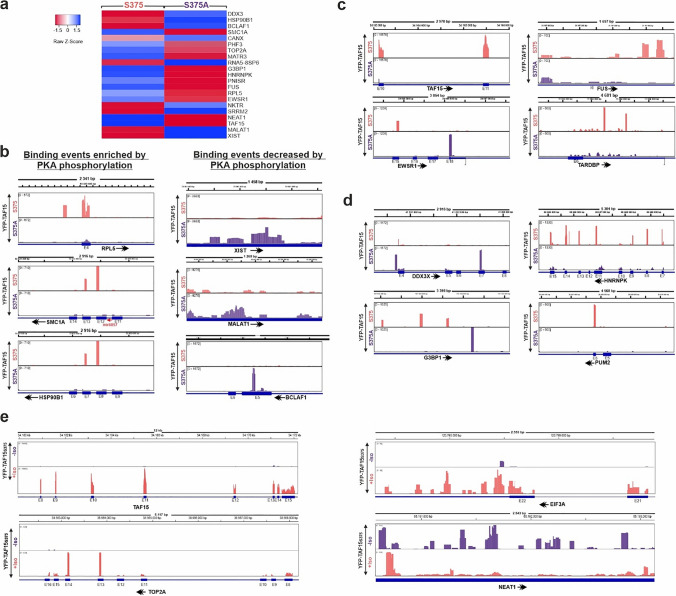
Fig. 3.
PKA phosphorylation affects RNA-binding patterns of TAF15. a Cluster analysis of the most-enriched TAF15 RNA targets of TAF15 S375 (wt) and S375A iCLIP libraries. Z-Scores were calculated and represented using the Heatmapper software [61]. b Genome distribution of several examples of TAF15 iCLIP RNA-binding events influenced by PKA phosphorylation. Read count distribution is shown from the TAF15 S375/S375A iCLIP libraries using the Integrated Genome Browser, IGB [46]. Genomic position, annotation and direction of the gene transcription are shown (E; exons, I; introns). Quantification of mapped read counts is shown in brackets in each case. c Genome distribution of the read counts from TAF15 S375/S375A iCLIP libraries of FET protein family members using the IGB. d Genome distribution of the read counts from TAF15 S375/S375A iCLIP libraries of RNA granule RBPs TAF15 targets influenced by PKA phosphorylation and visualized using the IGB. e Genome distribution of the TAF15 iCLIP RNA-binding events in S375 iCLIP libraries under non-cAMP/cAMP elevation by Isoproterenol treatment and visualized by IGB