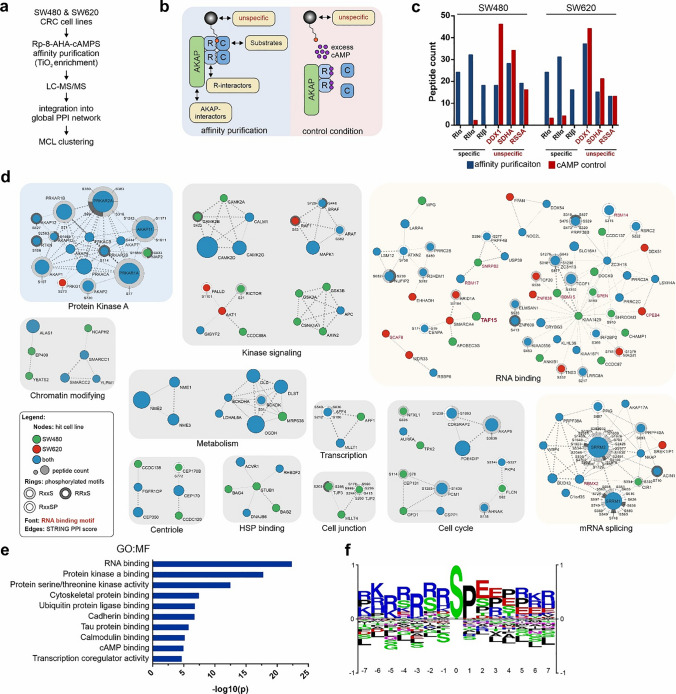
Fig. 4.
Macromolecular PKA complexes are enriched for RNA-binding proteins in CRC cell lines. a Workflow of the applied LC–MS and PPI network analysis. b Expected composition of macromolecular PKA complexes isolated via a for both, purification- and control condition (excess of cAMP). c Comparison of the two conditions for specific binders (highly abundant PKA subunits, black font) and unspecific binders (arbitrarily selected, red font) in both cell lines. d Network clusters obtained via workflow shown in a. Clusters were tested for functional enrichment and grouped accordingly. Node color indicates cell line specificity, node size indicates the number of identified peptides. Identified phosphorylation of sites matching the PKA consensus motif (RxxS in light grey, RRxS in dark grey and RxxSP in white) are indicated with rings encircling the nodes and labeled with the respective amino acid position. Red labels denote proteins containing RNA-binding domains. e Gene ontology (GO) analysis showing selected enriched molecular function terms of affinity isolated proteins. Analysis performed via STRING [41]. f Phosphomotif analysis of all detected phosphosites, generated via PhosphoSitePlus [67]