Abstract
Altered expressions of proto-oncogenes have been reported during normal lymphocytes mitogenesis and in T and B lymphocytes in patients with autoimmune diseases. We have recently demonstrated a significantly decreased expression of c-kit and c-Myc in NK cells isolated from patients with cancer, which might be related to the functional deficiency of NK cells in the tumor environment. Here, focusing on the regulatory mechanisms of this new clinical phenomenon, we determined expression of c-Myc, Notch1, Notch2, p-53, Cdk6, Rb and phosphorylated Rb in NK cells isolated from the healthy donors and cancer patients. The results of our study revealed a significant down-regulation of expression of Notch receptors and up-regulation of Cdk6 expression in NK cells in cancer, while no significant changes in the expression of p53 and Rb proteins were seen. These data revealed novel signaling pathways altered in NK cells in the tumor environment and support further investigation of the origin of deregulated expression of proto-oncogenes in NK cells patients with different types of cancer.
Keywords: NK cells, T cells, c-myc, Notch1, Notch2
Introduction
Cancer is a complex disease primarily driven by divergent signaling pathways of oncogenes and associated with the development of the tumor microenvironment. Oncogenes contribute to cancer development and diversity not only by inducing proliferation but also by developmental reprogramming of the epigenome [1]. Therefore, the tumor microenvironment is characterized by variable and dynamic patterns of cellular diversity, which leads to intratumoral heterogeneity, genetic and epigenetic diversification, growth dynamics and sensitivity to specific treatments.
Members of the Myc family of proto-oncogenes are the most commonly deregulated genes in human neoplasms. Myc proteins drive an increase in cellular proliferation and facilitate multiple aspects of tumor initiation and progression, thereby controlling all hallmarks of cancer [2]. For instance, c-Myc is a well-known transcription factor playing diverse roles in cell cycle progression, apoptosis and longevity during embryogenesis and oncogenesis; elevated expression of c-Myc is associated with aggressive carcinogenesis and a poor clinical outcome [3, 4]. However, involvement of oncogenes in cancer development is not limited to the transforming effect on malignant cells. Recently, Myc expression at the tumor site has been shown to regulate the tumor microenvironment through its effects on both innate and adaptive immune effector cells and immune regulatory pathways [5, 6]. Furthermore, Myc is suggested to be one of the essential transcriptional factors required for TCR-mediated signaling, T cell proliferation and survival, Th2 and Th17 differentiation [7, 8]. It regulates macrophage proliferation and metabolism [9], tolerogenic phenotype of dendritic cells [10] and germinal center B cell development and activation [11]. However, the function of immune cell oncogenes in tumor-bearing host is mostly unknown.
We have recently tested whether the c-Myc may be involved in the mechanism of NK cell abnormalities in patients with lung and gastric cancer by analyzing c-Myc mRNA and protein expression, MAPK and STAT3 activity, cell cycle and cell longevity in peripheral blood NK cells harvested from healthy donors and patients with cancer. Our results revealed a significantly decreased expression of c-Myc mRNA and protein, mitotic arrest of NK cells in different phases of cell cycle and a significant deregulation of NK cell death in cancer patients [12, 13]. These data allow speculating that defects of NK cell-mediated tumor surveillance may be associated with disturbed c-Myc expression in these cells in patients with cancer [14]. Interestingly, changes in proto-oncogene in NK cells in the tumor environment were different from those described in neoplastic cells. Proto-oncogenes themselves or suppressor genes that regulate them are often mutated in cancerous cells and this usually activates metabolic processes and enhances cell proliferation. But the defect of proto-oncogenes in NK cells was mostly associated with a decreased mRNA and protein expression. This raises the question about the origin of the proto-oncogene defects in NK cells in cancer. Marked phenotypic and functional alterations of NK cells in the tumor environment in vitro and in vivo have been repeatedly described. Alterations include changed decreased cytotoxicity and reduced IFN-γ production [12, 13]. Furthermore, c-Myc is involved in IL-15 signaling pathway, which is critical for NK cell maturation and homeostasis.
The deregulation of proto-oncogene Myc plays a key role in human oncogenesis, but in contrast to other proto-oncogenes, whose activity is due to mutation or truncation, Myc deregulation may be associated with the protein overexpression [15]. Potential mechanisms of c-Myc upregulation include gene amplification, regulation by the tissue-specific noncoding long-range enhancers and transcriptional upregulation [16]. For instance, one model states that the tumor suppressor p53 mediates transcriptional repression of c-Myc via a mechanism that involves binding to the c-myc promoter and elevation of histone deacetylation [17]. As p53 has been associated with the recruitment of histone deacetylases to its repressed gene targets [18], a similar repression mechanism may operate at the c-Myc promoter. Thus, the loss function of mutated p53 onco-repressor can lead to over-expression of c-Myc in cancer cells; although early reports suggest that this may involve transcriptional repression, new data suggest that p53-inducible miRNA(s) and subsequently repressed c-Myc are new players in p53/c-Myc regulatory network [19]. Interestingly, p53 has been reported to also regulate NK cell functional maturation [20]. Analysis of p53 expression at various stages of NK cell development revealed a lower level of p53 expression in less mature cell subsets compared to more mature cells. Thus, it is important to understand the functional significance of p53-dependent and p53-independent deregulation of c-Myc expression in NK cells seen in patients with cancer.
C-Myc functioning is regulated by several signal transduction pathways. For instance, c-Myc is one of the downstream targets of Notch signal transduction [21]. Notch1 can directly bind to the c-Myc promoter and signals from Notch receptors drive c-Myc-mediated cellular growth and metabolism. Importantly, Notch1 and Notch2, but not Notch3 and Notch 4, are expressed in human peripheral blood NK cells [22], and the Notch signaling pathway plays a substantial role in human NK cell development and function [23]. For instance, Notch ligands have been shown to have differential ability to induce and expand immature, but functional, NK cells from CD34+ human hematopoietic progenitor cells suggesting that Notch signaling is important for the physiologic development of NK cells at differentiation stages [24]. Activation of Notch1 in NK cells can also lead to an increased secretion of IFN-gamma production [22] demonstrating the importance of this signaling in NK cell function.
At the same time, involvement of c-Myc in NK cell proliferation has not been yet investigated, although we have reported a few correlational abnormalities in NK cell isolated from cancer patients including the cell cycle arrest of NK cells in the G0/G1 phase [12, 13]. In normal cells, Myc levels are nearly undetectable during quiescence, while Myc mRNA and proteins are rapidly induced upon cell cycle entry and remain relatively constant during cell cycle progression [25]. In fact, high expression of c-Myc leads to a shortened G1 phase, while reduced c-Myc expression causes lengthening of the cell cycle [26]. Cyclin-dependent kinase 6 (Cdk6) plays a vital role in regulating the progression of the cell cycle in part via controlling activity of tumor suppressor retinoblastoma protein Rb [27]. The absence of c-Myc during the G0-to-S phase transition can significantly reduce Cdk6 activity and result in delayed of Rb phosphorylation [28]. However, there are no data investigating a potential alteration of Rb and Cdk6 expression and activity associated with the mitotic arrest of NK cells in G0/G1 phase of the cell cycle reported in cancer patients.
These findings prompted us to ask which mechanisms may coordinate expression of proto-oncogene c-Myc in NK cells in cancer patients. Our studies show a significant downregulation of Notch1 receptor expression on peripheral blood NK cells in cancer patients with no significant changes in p53 expression or Rb activation in the same cells.
Materials and methods
Patients’ samples
Peripheral blood specimens were collected from 35 patients: 25 men and ten women [median age 68 (36–79)]. Twenty patients (13—men and 7—women) had lung cancer (adenocarcinoma—16 cases, squamous cell carcinoma—4 cases, including 3 patients with metastases in the mediastinum) and 15 patients were with cancer of the gastrointestinal tract including gastric cancer (all adenocarcinomas—9 cases) and esophageal cancer (adenocarcinoma—1 cases and squamous cell carcinoma—5 cases, 4 patients with metastases in the lymph nodes and lungs). Stages of lung cancer were: I–II—6 patients, III—14 patients, IV—1 patient. Stages of gastric cancer were: II—4 patients, III—10 patients, IV—1 patient. Blood specimens were collected prior to the surgical and chemotherapy procedures. Healthy controls [n = 17, seven men and ten women with median age 55 (46–67)] were recruited from the personnel of the laboratories of the Research Institute of Fundamental and Applied Medicine. All studies were approved by the Institutional Review Board and all patients and healthy donors provided signed written informed consent for sample acquisition for research purposes.
Purification of NK and CD3+T cells
PBMC were isolated from the peripheral blood by Ficoll-Paque PLUS (Life Technologies, USA) density gradient centrifugation (2-16KL centrifuge, Sigma, Germany). NK cells were negatively selected using DynaMag Magnet with Dynabeads Untouched Human NK Cells Isolation kit (Life Technologies). T cells were isolated from PBMC by positive selection using DynaMag Magnet with Dynabeads Untouched Human CD3 microbeads (Life Technologies). The purity of the cell subsets was confirmed by flow cytometry (FACSCanto II, BD Biosciences, USA) using specific CD56 and CD16 monoclonal antibodies conjugated with FITC and PerCP and specific CD3 monoclonal antibodies labeled with FITC (BD Biosciences, USA) as described earlier 15. Data analysis was performed with BD FACSDiva Software. The purity of isolated NK cells was typically around 95% and potential contamination with NKT cells was less than 1% according to the manufacturer procedure.
Detection of Notch1, Notch2, p53, Cdk6 and Rb protein expression
Notch1 and Notch2 transmembrane proteins were detected on the surface of isolated NK cells using APC-conjugated mouse anti-human Notch1 and PE-conjugated anti-human Notch2 monoclonal antibodies (BioLegend, USA). For the intracellular p53 and Cdk6 expression, isolated NK cells were fixed (BD Phosflow Fix Buffer I) for 10 min at 37 °C, then permeabilized (BD Phosflow Perm Buffer III) on ice for 30 min, and then stained with FITC-conjugated mouse monoclonal anti-human p53 antibody (BioLegend) or Cdk6 antibody (Santa Cruz Biotechnology, USA). Intracellular expression of the total and phosphorylated (pS807/pS811) Rb proteins in fixed and permeabilizated NK cells was assessed using FITC-conjugated mouse monoclonal anti-Rb (IF8, Santa Cruz Biotechnology) and PE-conjugated mouse anti-Rb (pS807/pS811) (Phosflow, BD Biosciences) antibodies, respectively, in the same cells. Positive cells were assessed by flow cytometry on FACSCantoTM II (BD Biosciences) and analyzed by BD FACSDiva Software.
SmartFlare RNA detection assay
Isolated CD3+ T cells were incubated in RPMI-1640 medium (Sigma-Aldrich, USA) supplemented with 10% FCS (Sigma-Aldrich) in 96-well flat bottom plates (Corning, USA) at 37 °C, 5% CO2 for 20 h in the presence of Smart Flare MYC, Human, Cyanine 5 according to the manufacturer’s instructions (Merck Millipore, USA). Control probes were: (i) Cellular uptake control; (ii) Scramble control for specificity and (iii) Housekeeping control—the 18s gene. Fluorescence was detected by flow cytometry on BD FACSCantoTM II.
Statistical analysis
For a single comparison of two groups, the Student’s t test was used after the evaluation of normality. If data distribution was not normal, Mann–Whitney rank-sum test was performed. For the comparison of multiple groups, analysis of variance (ANOVA) was applied. SigmaStat Software was used for data analysis (SyStat Software Inc., USA). For all statistical analyses, p < 0.05 was considered significant. All experiments were repeated at least two times. Data are presented as the mean ± SEM.
Results
Reduced Notch1 expression in NK cells in cancer patients
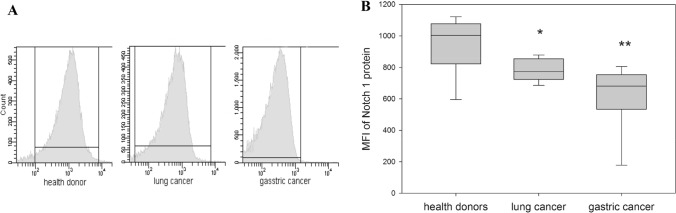
Expression of Notch1 transmembrane receptor was determined on the surface of the peripheral blood NK cells isolated from patients with lung and GI cancer and healthy donors (Fig. 1).
Fig. 1.
Differences in Notch1 expression in NK cells harvested from healthy donors and cancer patients. NK cells were isolated from the peripheral blood samples by negative selection using Dynabeads. The relative Notch1 expression was determined by flow cytometry on stained NK cells from healthy donors, patients with lung and gastric cancer. a The results of flow cytometry data from a representative experiment are shown. b Data of mean fluorescent intensity (MFI) are shown as the mean ± SEM (N = 11–20). *p < 0.05; **p < 0.01; one-way ANOVA
MFI data revealed that the expression of Notch1 protein on NK cells in patients with both lung cancer (803 ± 29) and gastric cancer (631 ± 65) was significantly reduced compared with the expression of Notch1 protein on NK cells from healthy volunteers (1001 ± 75) (p < 0.03 and p < 0.002, respectively). These results together with our previous data showing decreased expression of c-Myc in NK cells in patients with lung and gastric cancer [13] allow speculating about potential functional link between dysregulated Notch1 and c-Myc expression in NK cells in cancer. In fact, recent data identified c-Myc as an essential mediator of Notch1 signaling in lymphocytes and integrated Notch1 activation with oncogenic signaling pathways upstream of c-Myc [21, 29]. Figure 1 results also raised a question about Notch2 expression on the same NK cells.
Differential expression of Notch2 on NK cells in patients with lung and gastric cancer
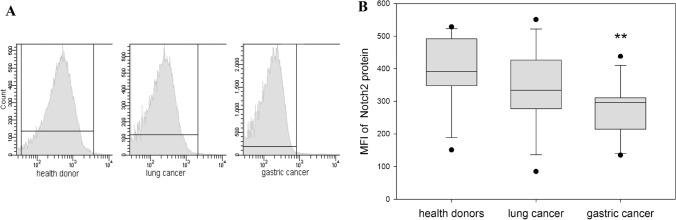
Expression of Notch2 transmembrane receptor was estimated on the surface of the same peripheral blood NK cells isolated from patients with lung cancer and gastric cancer in comparison to healthy donors (Fig. 2).
Fig. 2.
Differences in Notch2 expression in NK cells harvested from healthy donors and cancer patients. NK cells were isolated from the peripheral blood samples by negative selection using Dynabeads. Notch2 expression was determined by flow cytometry on stained isolated peripheral NK cells from healthy donors, patients with lung and gastric cancer. a The results of flow cytometry data from a representative experiment are shown. b Data of mean fluorescent intensity (MFI) are shown as the mean ± SEM (one-way ANOVA, N = 15–29). **p < 0.01
Unlike Notch1 expression, changes in Notch2 expression were not observed in all patients with cancer. NK cells obtained from patients with lung cancer expressed the similar levels of Notch2 (341 ± 33) as NK cells from healthy donors (389 ± 28) (p = 0.285). However, expression of Notch2 on NK cells in patients with gastric cancer was significantly reduced compared to the expression of Notch2 on NK cells from healthy donors (276 ± 23) (p < 0.005). This cancer-specific dysregulation of Notch2 expression on NK cells may suggest involvement of different pathways in Notch functioning and requires correlation with the expression or activation of other signaling molecules in NK cells.
Expression of p53, Cdk6, total Rb and pRb in NK cells from cancer patients and healthy donors
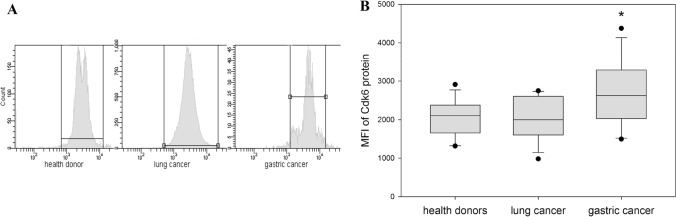
Our results revealed no statistically significant differences in p53 expression in NK cells between healthy donors (219 ± 17) and patients with lung (174 ± 19) and gastric cancer (208 ± 17, p = 0.107 and p = 0.679, respectively). Similarly, we did not reveal any significant differences in the expression of total and activated Rb in NK cells from healthy donors (Rb 808 ± 51 and pRb 6201 ± 726) and NK cells from patients with lung (Rb 737 ± 66 and pRb 4666 ± 499, p = 0.2 and p = 0.103, respectively) and gastric cancer (Rb 874 ± 87 and pRb 5544 ± 546, p = 0.26 and p = 0.481, respectively). However, expression of Cdk6 in NK cells demonstrated cancer-specific differences. It was significantly upregulated in NK cells from gastric cancer group (2676 ± 235, p = 0.030), but not in lung cancer group (2043 ± 158, p = 0.994) when compared with healthy volunteer results (2045 ± 132) (Fig. 3).
Fig. 3.
Differences in the levels of Cdk6 expression in NK cells harvested from healthy donors and cancer patients. NK cells were isolated from the peripheral blood samples by negative selection using Dynabeads. Cdk6 expression was determined by flow cytometry intracellularly in NK cells from healthy donors, patients with lung and gastric cancer. a The results of flow cytometry data from a representative experiment are shown. b Data of mean fluorescent intensity (MFI) are shown as the mean ± SEM (one-way ANOVA, N = 17–32). *p < 0.05
Reduced expression of c-Myc mRNA in CD3 T cells in cancer patients
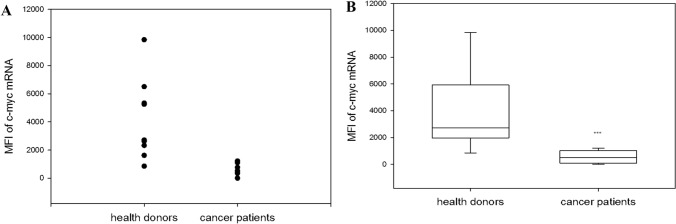
We have recently reported a reduced expression of c-Myc in NK cells obtained from patients with cancer [12, 13]. Because both T cells and NK cells are developed from a common lymphoid precursor, we next evaluated c-Myc mRNA in the general population of CD3 T cells obtained from cancer patients and healthy donors. Our results revealed a significant decrease in c-Myc mRNA expression in CD3+ T cells in cancer patients (6 patients with lung and 3 with gastric cancer). T cells in healthy donors expressed 4119 ± 953 c-myc mRNA and cancer patients—556 ± 160 (p = 0.001) (Fig. 4).
Fig. 4.
Differences in c-myc mRNA expression in T cells harvested from healthy donors and cancer patients. T cells were isolated from the peripheral blood samples by positive selection using BD IMag™ anti-human CD3 Particles DM, incubated in complete medium for 20 h. C-myc-mRNA expression in peripheral T cells from healthy donors and patients with lung and cancer was determined by Smart Flare. a Actual experimental results are shown. b Data of mean fluorescent intensity (MFI) are shown as the mean ± SEM (one-way ANOVA, N = 9). ***p = 0.001
These results suggest that dysregulation of c-Myc expression may originate from the early stages of lymphoid cell differentiation.
Discussion
The main goal of these studies was to continue exploring the mechanisms of abnormal expression and function of proto-oncogenes in human NK cells harvested from patients with different types of cancer. Based on our early data showing altered appearance of c-Myc in NK cells in cancer [12, 13] and the fact that Notch signaling is involved in NK cell development [23, 37], we hypothesized that a decrease in c-Myc expression might be associated with the changed Notch signaling in the same cells. This hypothesis was also supported by clinical data showing that Notch-induced signaling pathway accounts for the c-Myc expression in normal and malignant T cells [23, 30]. Our results here demonstrated that the expression of Notch1 receptor on NK cells was significantly down-regulated in all tested cancer patients (Fig. 1) regardless of the disease stage and the presence of metastases. Interestingly, Notch2 receptor expression was significantly reduced only in patients with gastric cancer when compared to healthy volunteers (Fig. 2). Thus, it is possible that a decrease in Notch1 expression, as well as in c-Myc expression, is a common feature of an immune abnormalities in NK cells seen in cancer patients, whereas a tumor-specific nature of Notch2 alterations in NK cells might suggest a more complex tissue-specific mechanism of NK cell regulation by Notch ligands. For instance, sustained Notch signaling can induce common lymphoid progenitor commitment to NK cell differentiation, although these NK cells might be immature in their cytokine production profile, hyporesponsive and poorly acquiring NK cell receptors involved in self-tolerance and effector function [31]. Further studies are needed to reveal mechanisms of abnormal expression of Notch receptors in NK cells in cancer and to directly prove their functional involvement in down-regulating c-Myc expression in the same NK cell subsets.
Relatively little is known concerning NK cell homeostasis, maintenance and turnover in patients with different forms of cancer. To gain more insight into NK cell dynamics, we have recently investigated the cell cycle status, turnover and apoptosis rates of NK cells in normal and cancerous conditions. We have reported an NK cell cycle arrest in the G0/G1 phase in cancer patients [13]. Therefore, here we have focused on potential alterations of the G1 cell cycle kinase Cdk6 as this kinase is a catalytic subunit of the protein kinase complex important for the G1-phase progression and G1/S transition of the cell cycle [32]. Interestingly, we found an increased expression of Cdk6 in NK cells isolated from patients with gastric, but not lung cancer when compared with the results seen in healthy donors (Fig. 3). Similar to the Notch2 expression (Fig. 2), alterations of Cdk6 in NK cells were cancer type-specific, but, contrary to the Notch2 expression, expression of Cdk6 was upregulated in NK cells from cancer patients. Although it was shown that the expression of Cdk6 could be Notch-dependent both in vitro and in vivo in human T cell acute lymphoblastic leukemia [33], nothing is known about Notch-associated regulation of G1 proteins in non-transformed NK cells in health and diseases. Our data provide a new model for investigating these crossed signaling pathways in immune cells in cancer.
Activation of cyclin-dependent kinases, including Cdk6, at specific time points during the cell cycle regulates phosphorylation and inactivation of the retinoblastoma protein. Based on the data suggesting the possibility that the premature and prolonged enhancement of Cdk activity in NK cells may be associated with, and therefore may contribute to, the reduced expression and phosphorylation of Rb, and the associated cell cycle arrest [34], our next task here was to assess a potential involvement of activated Rb in the abnormal NK cell cycling in cancer. However, no significant changes in the expression of the total and phosphorylated Rb in NK cells isolated from cancer patients were revealed. Altogether our results allowed concluding that NK cell cycle arrest in cancer patients in the G0/G1 phase is associated with the reduced c-Myc expression and mediated by Notch signaling and Cdk6 function, while likely being p53- and Rb-independent. In fact, previous studies revealed the oncogenic potential of aberrant Notch signaling in primary and transformed lymphocytes through its deregulation of the cell cycle [33].
In agreement with this, our studies also suggest that the defects of NK cells in cancer patients may be associated with an uncharacterized disturbance of the hematopoietic progenitor cells, in particular, the common lymphoid precursors. In cancer, the failure of T lymphocytes, similarly to NK cells, to eradicate transformed cells is mainly attributed to the dysfunctional hyporesponsive phenotype [35]. T cell exhaustion and senescence induced in the tumor environment are central dysfunctional states of immune effectors found in cancer patients, which hamper active antitumor immunity and immunotherapy and sustain the suppressive properties of the tumor microenvironment [36]. Importantly, several observations suggest that exhausted T cells acquire epigenetic modifications that are distinguishable from those of effector T cells [37]. Here, we tested whether c-Myc expression in T cells from cancer patients could be down-regulated in a manner similar to the expression in NK cells from the same patients. As shown in Fig. 4, c-Myc expression in T cells harvested from patients with lung and gastric cancers was significantly reduced when compared with the results obtained in healthy donors.
Comparative decrease in c-Myc expression in both NK and T cells in cancer might suggest a wider distribution of c-Myc dysregulation in immune cells, for instance, in common lymphoid precursors. Though this hypothesis requires experimental verification, it is well supported by emerging evidence indicating that cancer induces alterations of the myelopoietic output, defined as emergency myelopoiesis [38]. This pathway, which leads to the generation of different myeloid populations endowed with tumor-supporting activities, is the result of a multistep process encompassing initial events originating into the bone marrow [39]. For instance, cancer-induced abnormalities in differentiation and function of early-stage committed unipotent and lineage-specific neutrophils, monocytes, immature myeloid progenitors in the bone marrow have been repeatedly reported [40–43]. While normal hematopoiesis requires the precise orchestration of lineage-specific patterns of gene expression at each stage of development via epigenetic regulators, disordered epigenetic regulation has emerged as a key mechanism contributing not only to hematological malignancies, but also to solid tumors. Epigenetic mechanisms, which involve DNA modification, covalent histone modification and RNA interference, result in the heritable downregulation or silencing of gene expression without a change in DNA sequences. Epigenetic modification of myeloid precursor functional plasticity leads to the remodeling of its characteristics, therefore reframing the microenvironment towards countering tumor growth and metastasis [44]. Similar mechanisms can be suggested for the lymphopoiesis in cancer, including NK cell development, but direct evidence is still missing. Understanding the biology and behavior of NK cell generation, differentiation and deregulation in the tumor environment should allow the development of new therapeutic targets for a variety of NK cell-related diseases, including cancer.
Acknowledgements
We are grateful to Sergey Bernikov for great help in formal analysis, application of statistical, mathematical, computational and other techniques to analyze or synthesize the study data.
Author contribution
Conceptualization, writing, original draft preparation, GKZ; methodology, project administration, supervision, NNN; formal analysis, investigation, data curation, EK, NTU, GVS; resources, ETB, VAM, GMT; writing, review and editing, ZMB; writing, review and editing, visualization, funding acquisition, MRS.
Funding
This research was funded by the Committee on science of the Ministry of Education and Science of the Republic of Kazakhstan (№AP05130612).
Compliance with ethical standards
Conflict of interest
The authors declare no potential conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Gulnur K. Zakiryanova, Email: gzakiryanova@gmail.com
Michael R. Shurin, Email: shurinmr@upmc.edu
References
- 1.Vicente-Duenas C, Romero-Camarero I, Cobaleda C, Sanchez-Garcia I. Function of oncogenes in cancer development: a changing paradigm. EMBO J. 2013;32:1502–1513. doi: 10.1038/emboj.2013.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Venkateswaran N, Conacci-Sorrell M. MYC leads the way. Small GTPases. 2017;11(2):86–94. doi: 10.1080/21541248.2017.1364821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hofmann JW, Zhao X, De Cecco M, Peterson AL, Pagliaroli L, Manivannan J, Hubbard GB, Ikeno Y, Zhang Y, Feng B, Li X, Serre T, Qi W, Van Remmen H, Miller RA, Bath KG, de Cabo R, Xu H, Neretti N, Sedivy JM. Reduced expression of MYC increases longevity and enhances healthspan. Cell. 2015;160:477–488. doi: 10.1016/j.cell.2014.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Conacci-Sorrell M, McFerrin L, Eisenman RN. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Casey SC, Tong L, Li Y, Do R, Walz S, Fitzgerald KN, Gouw AM, Baylot V, Gutgemann I, Eilers M, Felsher DW. MYC regulates the antitumor immune response through CD47 and PD-L1. Science. 2016;352:227–231. doi: 10.1126/science.aac9935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casey SC, Baylot V, Felsher DW. The MYC oncogene is a global regulator of the immune response. Blood. 2018;131:2007–2015. doi: 10.1182/blood-2017-11-742577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preston GC, Sinclair LV, Kaskar A, Hukelmann JL, Navarro MN, Ferrero I, MacDonald HR, Cowling VH, Cantrell DA. Single cell tuning of Myc expression by antigen receptor signal strength and interleukin-2 in T lymphocytes. EMBO J. 2015;34:2008–2024. doi: 10.15252/embj.201490252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gnanaprakasam JN, Wang R. MYC in regulating immunity: metabolism and beyond. Genes (Basel) 2017;8(3):88. doi: 10.3390/genes8030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu L, Lu Y, Martinez J, Bi Y, Lian G, Wang T, Milasta S, Wang J, Yang M, Liu G, Green DR, Wang R. Proinflammatory signal suppresses proliferation and shifts macrophage metabolism from Myc-dependent to HIF1alpha-dependent. Proc Natl Acad Sci U S A. 2016;113:1564–1569. doi: 10.1073/pnas.1518000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Gonzalez PA, Maggi J, Schinnerling K, Sepulveda-Gutierrez A, Soto L, Neira O, Mehdi AM, Nel HJ, Pesce B, Aravena O, Molina MC, Catalan D, Thomas R, Verdugo RA, Aguillon JC. Regulation of tolerogenic features on dexamethasone-modulated MPLA-activated dendritic cells by MYC. Front Immunol. 2019;10:1171. doi: 10.3389/fimmu.2019.01171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robaina MC, Mazzoccoli L, Klumb CE. Germinal centre B cell functions and lymphomagenesis: circuits involving MYC and MicroRNAs. Cells. 2019;8(11):1365. doi: 10.3390/cells8111365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zakiryanova GK, Kustova E, Urazalieva NT, Amirbekov A, Baimuchametov ET, Nakisbekov NN, Shurin MR. Alterations of oncogenes expression in NK cells in patients with cancer. Immun Inflamm Dis. 2017;5:493–502. doi: 10.1002/iid3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zakiryanova GK, Kustova E, Urazalieva NT, Baimuchametov ET, Nakisbekov NN, Shurin MR. Abnormal expression of c-Myc oncogene in NK cells in patients with cancer. Int J Mol Sci. 2019;20(3):756. doi: 10.3390/ijms20030756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zakiryanova GK, Wheeler S, Shurin MR. Oncogenes in immune cells as potential therapeutic targets. Immunotargets Ther. 2018;7:21–28. doi: 10.2147/ITT.S150586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med. 2014;4(6):a014241. doi: 10.1101/cshperspect.a014241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lancho O, Herranz D. The MYC enhancer-ome: long-range transcriptional regulation of MYC in cancer. Trends Cancer. 2018;4:810–822. doi: 10.1016/j.trecan.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho JS, Ma W, Mao DY, Benchimol S. p53-Dependent transcriptional repression of c-myc is required for G1 cell cycle arrest. Mol Cell Biol. 2005;25:7423–7431. doi: 10.1128/MCB.25.17.7423-7431.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 19.Sachdeva M, Mo YY. p53 and c-myc: how does the cell balance "yin" and "yang"? Cell Cycle. 2009;8:1303. doi: 10.4161/cc.8.9.8362. [DOI] [PubMed] [Google Scholar]
- 20.Collin R, St-Pierre C, Guilbault L, Mullins-Dansereau V, Policheni A, Guimont-Desrochers F, Pelletier AN, Gray DH, Drobetsky E, Perreault C, Hillhouse EE, Lesage S. An unbiased linkage approach reveals that the p53 pathway is coupled to NK cell maturation. J Immunol. 2017;199:1490–1504. doi: 10.4049/jimmunol.1600789. [DOI] [PubMed] [Google Scholar]
- 21.Palomero T, Lim WK, Odom DT, Sulis ML, Real PJ, Margolin A, Barnes KC, O'Neil J, Neuberg D, Weng AP, Aster JC, Sigaux F, Soulier J, Look AT, Young RA, Califano A, Ferrando AA. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc Natl Acad Sci U S A. 2006;103:18261–18266. doi: 10.1073/pnas.0606108103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manaster I, Gazit R, Goldman-Wohl D, Stern-Ginossar N, Mizrahi S, Yagel S, Mandelboim O. Notch activation enhances IFN-gamma secretion by human peripheral blood and decidual NK cells. J Reprod Immunol. 2010;84:1–7. doi: 10.1016/j.jri.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 23.Felices M, Ankarlo DE, Lenvik TR, Nelson HH, Blazar BR, Verneris MR, Miller JS. Notch signaling at later stages of NK cell development enhances KIR expression and functional maturation. J Immunol. 2014;193:3344–3354. doi: 10.4049/jimmunol.1400534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck RC, Padival M, Yeh D, Ralston J, Cooke KR, Lowe JB. The Notch ligands Jagged2, Delta1, and Delta4 induce differentiation and expansion of functional human NK cells from CD34+ cord blood hematopoietic progenitor cells. Biol Blood Marrow Transplant. 2009;15:1026–1037. doi: 10.1016/j.bbmt.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hann SR, Thompson CB, Eisenman RN. c-myc oncogene protein synthesis is independent of the cell cycle in human and avian cells. Nature. 1985;314:366–369. doi: 10.1038/314366a0. [DOI] [PubMed] [Google Scholar]
- 26.Shichiri M, Hanson KD, Sedivy JM. Effects of c-myc expression on proliferation, quiescence, and the G0 to G1 transition in nontransformed cells. Cell Growth Differ. 1993;4:93–104. [PubMed] [Google Scholar]
- 27.Malumbres M, Barbacid M. Mammalian cyclin-dependent kinases. Trends Biochem Sci. 2005;30:630–641. doi: 10.1016/j.tibs.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 28.Mateyak MK, Obaya AJ, Sedivy JM. c-Myc regulates cyclin D-Cdk4 and -Cdk6 activity but affects cell cycle progression at multiple independent points. Mol Cell Biol. 1999;19:4672–4683. doi: 10.1128/MCB.19.7.4672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, Li Y, Wolfe MS, Shachaf C, Felsher D, Blacklow SC, Pear WS, Aster JC. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rolink AG, Balciunaite G, Demoliere C, Ceredig R. The potential involvement of Notch signaling in NK cell development. Immunol Lett. 2006;107:50–57. doi: 10.1016/j.imlet.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Bachanova V, McCullar V, Lenvik T, Wangen R, Peterson KA, Ankarlo DE, Panoskaltsis-Mortari A, Wagner JE, Miller JS. Activated notch supports development of cytokine producing NK cells which are hyporesponsive and fail to acquire NK cell effector functions. Biol Blood Marrow Transplant. 2009;15:183–194. doi: 10.1016/j.bbmt.2008.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tigan AS, Bellutti F, Kollmann K, Tebb G, Sexl V. CDK6-a review of the past and a glimpse into the future: from cell-cycle control to transcriptional regulation. Oncogene. 2016;35:3083–3091. doi: 10.1038/onc.2015.407. [DOI] [PubMed] [Google Scholar]
- 33.Joshi I, Minter LM, Telfer J, Demarest RM, Capobianco AJ, Aster JC, Sicinski P, Fauq A, Golde TE, Osborne BA. Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood. 2009;113:1689–1698. doi: 10.1182/blood-2008-03-147967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamauchi A, Bloom ET. Control of cell cycle progression in human natural killer cells through redox regulation of expression and phosphorylation of retinoblastoma gene product protein. Blood. 1997;89:4092–4099. doi: 10.1182/blood.V89.11.4092. [DOI] [PubMed] [Google Scholar]
- 35.Thommen DS, Schumacher TN. T cell dysfunction in cancer. Cancer Cell. 2018;33:547–562. doi: 10.1016/j.ccell.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao Y, Shao Q, Peng G. Exhaustion and senescence: two crucial dysfunctional states of T cells in the tumor microenvironment. Cell Mol Immunol. 2020;17:27–35. doi: 10.1038/s41423-019-0344-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, Tonnerre P, Chung RT, Tully DC, Allen TM, Frahm N, Lauer GM, Wherry EJ, Yosef N, Haining WN. The epigenetic landscape of T cell exhaustion. Science. 2016;354:1165–1169. doi: 10.1126/science.aae0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ueha S, Shand FH, Matsushima K. Myeloid cell population dynamics in healthy and tumor-bearing mice. Int Immunopharmacol. 2011;11:783–788. doi: 10.1016/j.intimp.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Sica A, Guarneri V, Gennari A. Myelopoiesis, metabolism and therapy: a crucial crossroads in cancer progression. Cell Stress. 2019;3:284–294. doi: 10.15698/cst2019.09.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Richards DM, Hettinger J, Feuerer M. Monocytes and macrophages in cancer: development and functions. Cancer Microenviron. 2013;6:179–191. doi: 10.1007/s12307-012-0123-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu YP, Padgett L, Dinh HQ, Marcovecchio P, Blatchley A, Wu R, Ehinger E, Kim C, Mikulski Z, Seumois G, Madrigal A, Vijayanand P, Hedrick CC. Identification of an early unipotent neutrophil progenitor with pro-tumoral activity in mouse and human bone marrow. Cell Rep. 2018;24:2329–2341.e2328. doi: 10.1016/j.celrep.2018.07.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casbon A-J, Reynaud D, Park C, Khuc E, Gan DD, Schepers K, Passegué E, Werb Z. Invasive breast cancer reprograms early myeloid differentiation in the bone marrow to generate immunosuppressive neutrophils. Proc Natl Acad Sci U S A. 2015;112:E566–E575. doi: 10.1073/pnas.1424927112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lecot P, Sarabi M, Pereira Abrantes M, Mussard J, Koenderman L, Caux C, Bendriss-Vermare N, Michallet MC. Neutrophil heterogeneity in cancer: from biology to therapies. Front Immunol. 2019;10:2155. doi: 10.3389/fimmu.2019.02155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C, Wang S, Liu Y, Yang C. Epigenetics in myeloid derived suppressor cells: a sheathed sword towards cancer. Oncotarget. 2016;7:57452–57463. doi: 10.18632/oncotarget.10767. [DOI] [PMC free article] [PubMed] [Google Scholar]