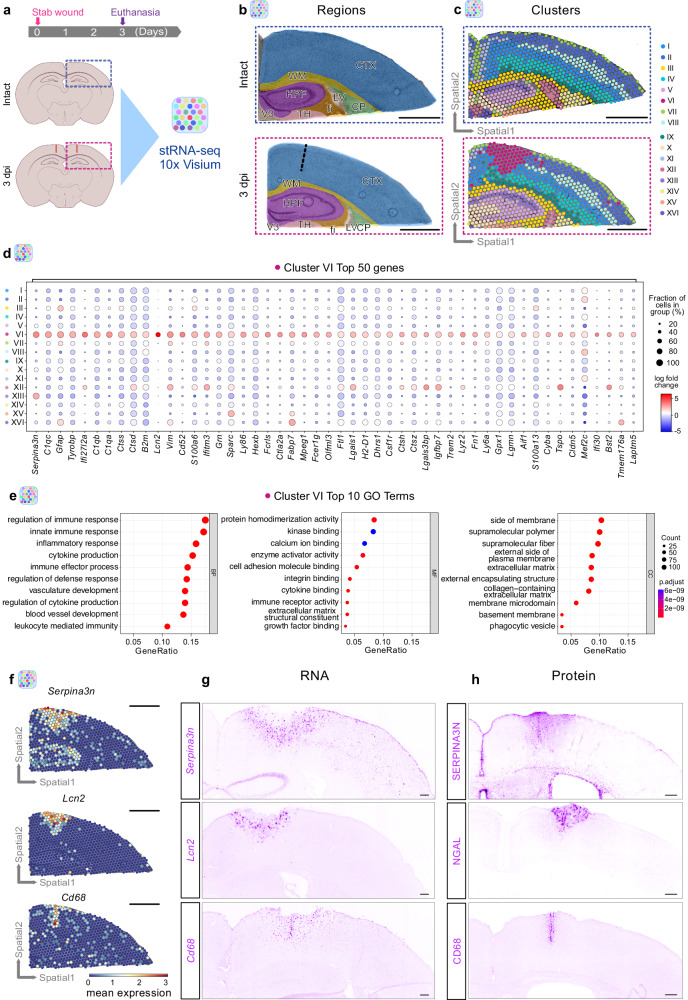
Fig. 1. Spatially resolved transcriptomic changes induced by stab wound injury.
a Experimental scheme to conduct spatial transcriptomics in intact and stab wound-injured mouse cerebral cortices (3 dpi). Brains were manually resected, and selected areas highlighted in blue or red dashed boxes were positioned on 10x Visium capture areas. b Brain sections of both conditions contain cortical, hippocampal, and white matter regions. The black dashed line indicates the injury core. c Clustering of gene expression data on spatial coordinates based on highly variable genes and subsequent dimensionality reduction. d Dot plot illustrating the expression of the 50 most enriched genes in the injury-induced cluster VI. e Dot plots depicting GO terms over-represented in cluster VI significantly enriched genes (pval < 0.05, log2 fold change >1, method t-test overestimated_variance). Significance of GO terms was determined by performing GO enrichment analysis on gene sets. Spatial transcriptomic analysis (a–e) is based on n = 1 animal per condition over 1 experiment, with two sections being captured. f Gene expression of cluster VI-enriched genes Serpina3n, Lcn2, and Cd68 in spatial context. g, h Images depicting expression of Serpina3n, Lcn2, and Cd68 at the RNA (g) and protein level (h) in stab wound-injured cerebral cortices at 3 dpi (n = 3 animals). All images are full z-projections of confocal z-stacks. Scale bars: b, c, f: 1 mm, g, h: 150 μm. stRNA-seq spatial transcriptomics, CTX cerebral cortex, WM white matter, HPF hippocampal formation, LV lateral ventricle, CP caudoputamen, V3 third ventricle, TH thalamus, fi fimbria, dpi days post-injury, BP biological processes, MF molecular functions, CC cellular components, GO gene ontology.