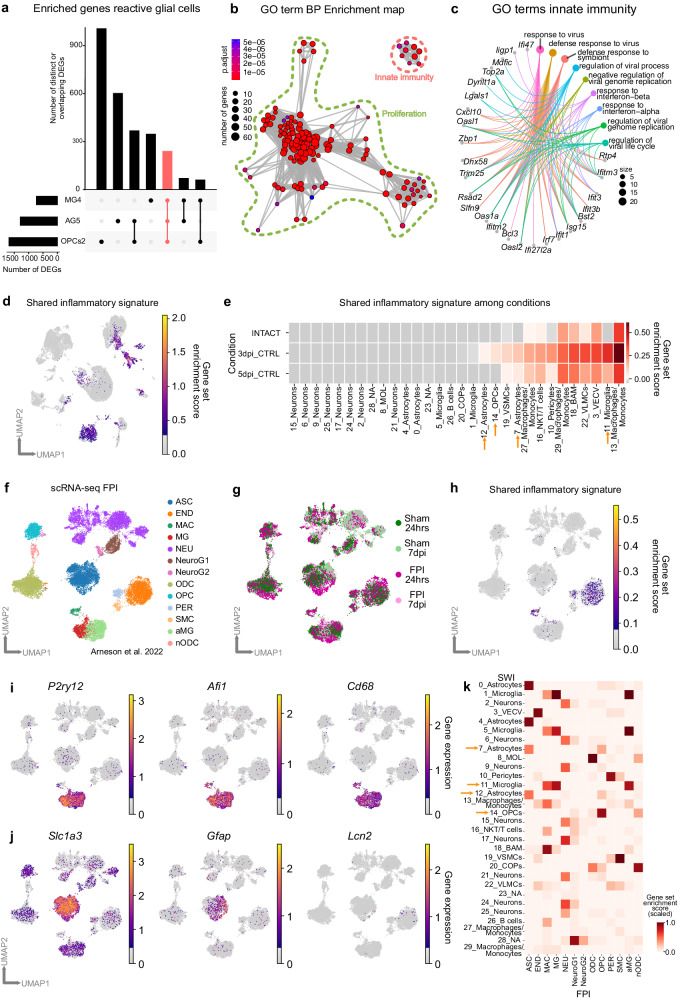
Fig. 5. Stab wound injury induces common transcriptional changes in reactive glial subpopulations characterizing a specific reactive state.
a UpSet plot depicting unique (single points) or overlapping (connected points) DEGs (pval < 0.05, log2 fold change >1.6, method t-test overestimated_variance). Vertical bars indicate the number of unique or shared genes between clusters. b GO term network analysis of the 241 commonly shared genes, associating shared DEGs with the biological processes of proliferation (green dashed line) and innate immunity (orange dashed line). Significance determined by method GO enrichment analysis on gene sets. c Chord diagram illustrating innate immunity GO terms from panel (b) and the corresponding genes. d UMAP plot depicting localization of the gene set score related to the shared inflammatory signature shown in panel (c) among all cell clusters from Fig. 4b. e Heatmap depicting the shared inflammatory signature gene set score. Data shown in panels (a)–(e) are derived from n = 12 intact animals over 3 independent experiments (5 scRNAseq libraries), n = 3 3dpi animals over 1 experiment (2 scRNAseq libraries), and n = 9 5dpi animals over 3 independent experiments (3 scRNAseq libraries). f UMAP plot illustrating single cells distributed among 13 distinct clusters in the FPI dataset. Clusters are annotated according to Arneson et al.50. g UMAP plot depicting the distribution of cells isolated from Sham (green) and FPI (purple) cerebral cortices at 24 h or 7 dpi. h UMAP plot depicting the localization of the gene set score related to the shared inflammatory signature from panel (c). i, j UMAPs highlighting expression of example marker genes to identify reactive microglial (i) and astrocytic clusters (j). k Heatmap displaying cluster similarity of SWI (y-axis) and FPI datasets (x-axis). Orange arrows highlighting clusters of interest. DEGs differentially expressed genes, GO gene ontology, BP biological processes, UMAP uniform manifold approximation and projection, CTRL stab wound-injured control animals, dpi days post-injury, COPs committed oligodendrocyte progenitors, MOL mature oligodendrocytes, VECV vascular endothelial cells (venous), VSMCs vascular smooth muscle cells, VLMCs vascular and leptomeningeal cells, BAM border-associated macrophages, NKT cells natural killer T cells, NA not available, ASC astrocytes, END endothelial, MAC macrophages, MG microglia, NEU neurons, NeuroG1 neurogenesis1, NeuroG2 neurogenesis2, ODC oligodendrocytes, OPC oligodendrocyte precursor, PER pericytes, SMC smooth muscle cells, aMG activated microglia, nODC new oligodendrocytes, hrs hours, FPI fluid percussion injury, SWI stab wound injury.