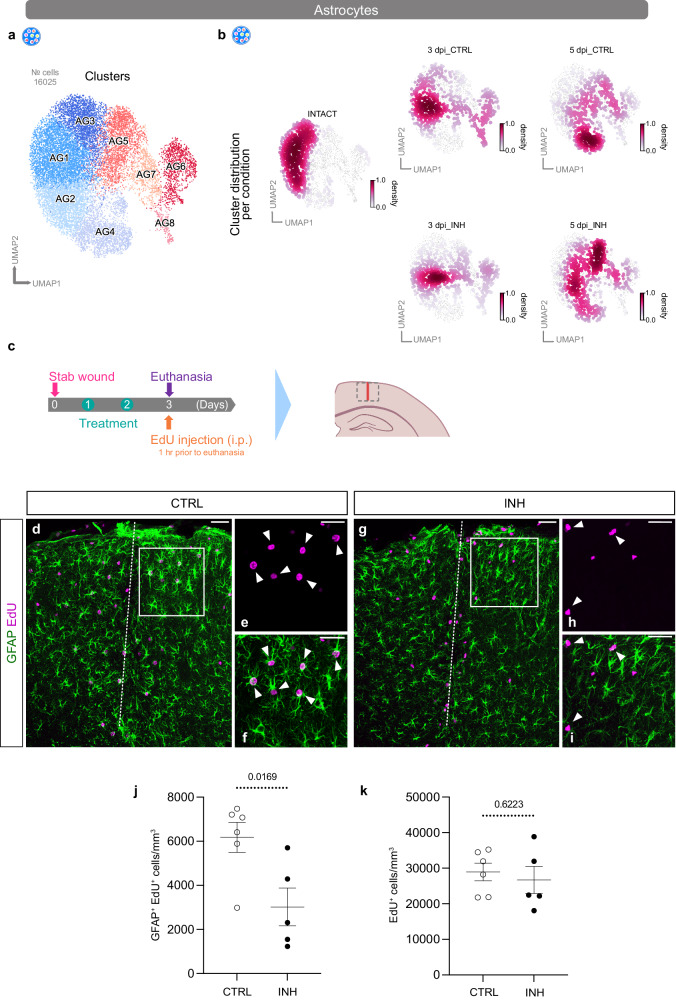
Fig. 8. Proliferation of reactive astrocytes is decreased after Cxcr3 and Tlr1/2 pathway inhibition.
a, b UMAPs illustrating subclusters of astrocytes (a) and cell distributions (b) among those subclusters at all time points (intact, 3 dpi, and 5 dpi) and conditions (CTRL and INH). Data in (b) are downsampled to an equal number of cells between time points and conditions. Data are derived from n = 12 intact animals over 3 independent experiments (5 scRNAseq libraries), n = 3 3dpi CTRL animals over 1 experiment (2 scRNAseq libraries), n = 3 3dpi INH animals over 1 experiment (2 scRNAseq libraries), n = 9 5dpi CTRL animals over 3 independent experiments (3 scRNAseq libraries) and n = 6 5dpi INH animals over 2 independent experiments (3 scRNAseq libraries). c Experimental paradigm for assessing astrocyte proliferation. The dashed gray box on mouse brain scheme indicates the analyzed area. The red line indicates the injury core. d, e Representative overview images of proliferating GFAP+ (green) and EdU+ (magenta) astrocytes in CTRL (d) and INH-treated (e) animals. White dashed lines highlight injury cores. Micrographs (e, f, h, i) are magnifications of white boxed areas in (d) and (g), respectively. White arrowheads in micrographs indicate colocalization of EdU (e, h) with GFAP+ astrocytes (f, i). All images are full z-projections of confocal z-stacks. j, k Dot plots depicting density of proliferating (GFAP+ and EdU+) astrocytes (j) and total density of proliferating (EdU+) cells (k) in CTRL and INH-treated animals. Data are shown as mean ± standard error of the mean. Each data point represents one animal. Source data are provided as Source Data file. Statistics in (j) and (k) have been derived from nCTRL = 6 and nINH = 5 animals. p-values were determined with unpaired t-test (two-tailed). Scale bars: d, g: 50 μm, e, i: 20 μm. UMAP uniform manifold approximation and projection, dpi days post-injury, EdU 5-ethinyl-2′-deoxyuridine, i.p. intraperitoneal injection, CTRL stab wound-injured control animals, INH stab wound-injured inhibitor-treated animals.