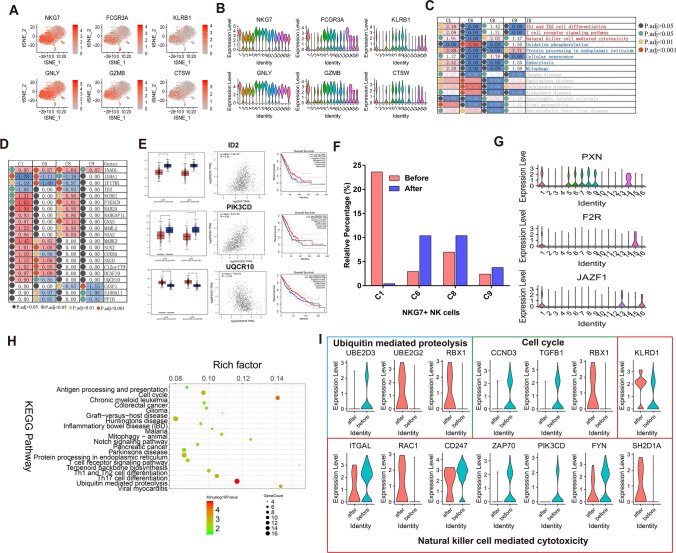
Fig. 4.
NKG7+ NK cells heterogeneity and molecular alterations after Pembrolizumab treatment. a, b T-SNE and violin plot of six common cell markers in NK cells (cluster 1, 6, 8, 9). c Shared significant enriched Pembrolizumab-responsive pathways in NK cells (cluster 1, 6, 8, 9). d Shared significant enriched Pembrolizumab-responsive genes in NK cells (cluster 1, 6, 8, 9). e Differential expression analysis, Pearson correlation analysis and overall survival analysis of differentially expressed genes (ID2, PIK3CD and UQCR10) using TCGA database. LUAD: lung adenocarcinoma, LUSC: lung squamous cell carcinoma. *p < 0.05. f The relative percentage of cluster 1, 6, 8, 9 before and after treatment with Pembrolizumab. g Violin plot showing the expression distribution of cluster 1 markers PXN, F2R and JAZF1. h Top 20 significant enriched pathways of differentially expressed genes in cluster 1. i Violin plot showing the most significantly altered genes involved in ubiquitin-mediated proteolysis, cell cycle and natural killer cell-mediated cytotoxicity, after Pembrolizumab treatment. Red represents after treatment with Pembrolizumab and blue represents before treatment with Pembrolizumab