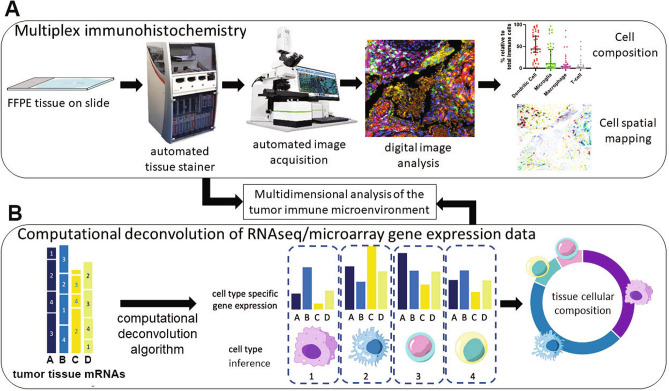
Fig. 1.
Mapping of the tumor immune microenvironment. a Multiplex Immunohistochemistry (mIHC). The process flow for multiplex immunohistochemical staining and analysis of tumor tissue using Opal™ technology. Formalin-fixed paraffin-embedded (FFPE) tissue is stained in an automated tissue processor and stainer, where multiple predetermined primary antibodies are used in a series of incubation and washing steps, followed by signal amplification. The slides are scanned on a multispectral microscope and images are acquired. The images are then analyzed using morphometric software to enumerate specific cell types and spatial proximity analysis to determine the physical location in two dimensions and the location of any cell type with any other cell type or histopathological biomarker. b Computational deconvolution applied to gene expression data. Illustration of the deconvolution of heterogeneous mRNAs derived from tumor tissue expression analysis, typically RNA-sequencing. From the left, gene expression of genes A, B, C and D represents all mRNAs expressed in the tumor bulk, including all cell types 1, 2, 3, 4. Following computational deconvolution, the cell types present in the tumor sample can be inferred and the proportion of each cell type in the tissue can be estimated, based on the input of cell gene expression matrices from previous experiments (e.g., single-cell mRNA sequencing). Data from mIHC analysis and computational deconvolution can be combined to map the cellular content and physical distribution of cells in the tumor microenvironment