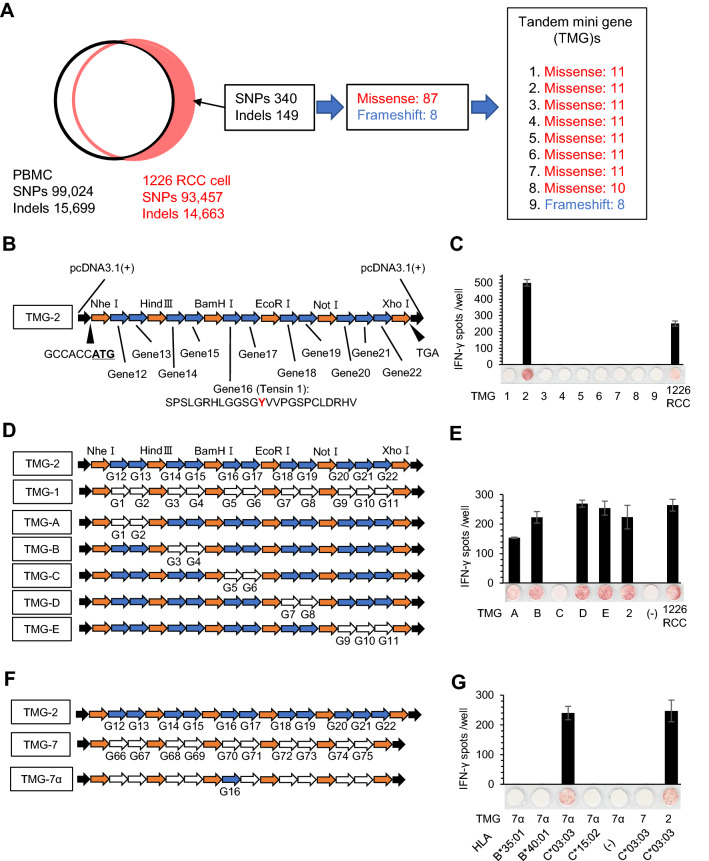
Fig. 3.
Identification of the tumor antigen recognized by the Q1 TIL clone. a Schematic summary of whole exome sequencing of 1226 RCC cells and design of TMGs. A total of 340 SNPs and 149 indel mutations were detected in the genome of 1226 RCC cells compared with the genome from the patient’s PBMCs. The mutations encoded 87 missense mutations and 8 frameshift mutations. The mutations were designed into 9 TMGs. b Design of TMGs. The gene started with the Kozak consensus sequence followed by a start codon. For missense mutations, TMGs encoded polypeptides containing mutated amino acid residues flanked on their N- and C-termini with 12 amino acids across restriction enzyme sites. TMGs were cloned into the pcDNA3.1 plasmid. For frameshift mutations, the TMGs encoded polypeptides of frameshifted amino acid residues flanked on their N-terminus with 12 amino acids. c The Q1 TIL clone recognizes TMG-2. Each TMG was transiently transfected into 293 T-C*03:03 cells and the reactivity of the Q1 TIL clone was assessed by an IFNγ ELISPOT assay. 1226 RCC cells were used as a positive control. IFNγ spots are shown as mean ± SEM (n = 3). d, e, f, g. The Q1 TIL clone recognizes gene 16 in an HLA-C*03:03-restricted manner. To narrow down the candidate gene, several TMGs were constructed (D and F). The reactivity of the Q1 TIL clone was assessed by an IFNγ ELISPOT assay (E and G). 1226 RCC cells were used as a positive control. IFNγ spots are shown as mean ± SEM (n = 3), and p values were calculated using a paired t test; *p < 0.01