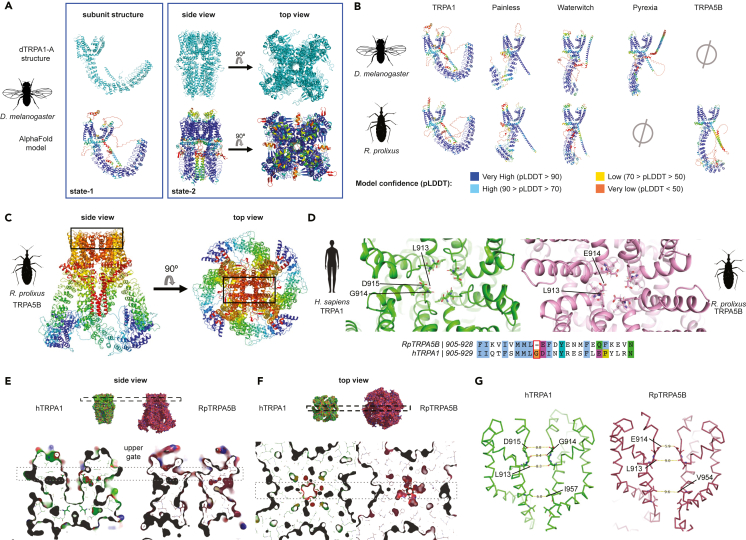
Figure 3.
Monomeric and tetrameric assemblies of RpTRPA5B channels modeled using AlphaFold, after validation with dTRPA1 structure
(A–D) (A) (left panel) Upper row, cartoon representation of chain A in tetramer of dTRPA1 in state 1 (PDB: 7YKR). The fold of a monomer in the experimentally determined structure of the dTRPA1 tetramer is very similar to the fold of an AlphaFold model of a single dTRPA1 monomer (bottom row). AlphaFold model colored from red to blue according to pLDDT confidence scores as shown in (B). The low-confidence regions (red) are not resolved in the reported structure and are likely to be intrinsically disordered. (Right panel) Upper row, experimentally determined structure of dTRPA1 in state 2 (PDB: 7YKS). Bottom row, tetrameric AlphaFold model of dTRPA1 depicted as cartoons colored from red to blue according to confidence scores as in (B). The N- and C-terminal regions, which are not resolved in 7YKS, were excluded in the prediction. Only the last five of the 17 ankyrin repeats (AR12-16) are visible in the structure and overall regions with low confidence in the model (red-yellow) are not resolved in the structure. (B) Monomers of Drosophila and Rhodnius TRPAs colored by pLDDT score from the AlphaFold modeling. (C) Tetrameric model of RpTRPA5B, colored as chain bows (N terminus, blue; C terminus, red). The black box indicates the location of the pore and selectivity filter shown in (D). (D) Top view of the selectivity filter of the pore of hTRPA1 (left: human TRPA1, PDB: 6V9Y) and RpTRPA5B (right). Three important residues61 – L913, G914, and D915, are marked in hTRPA1. The equivalent residues L913 and E914 are marked in RpTRPA5B, and G914 absent in RpTRPA5B is highlighted in the sequence alignment, together with additional residue changes adjacent to the selectivity filter.
(E and F) Comparison of the pore in hTRPA1 and model of RpTRPA5B indicates a closed upper gate and an open lower gate in the RpTRPA5B model. E. (upper row) Surface representation of hTRPA1 (green; PDB: 6V9Y) and RpTRPA5B (pink), side view. The dashed box indicates the location of the upper gate toward the outside of the cell. (lower row) Slab along the pore through the transmembrane domain of the hTRPA1 structure and RpTRPA5B model. F. (upper row) Top view of hTRPA1 and RpTRPA5B shown in E. (lower row) The slab is perpendicular to the pore at the level of the upper gate shown in E. The dashed box indicates the location of slab in (E).
(G) Distances between corresponding residues in the upper and lower gate in structures of hTRPA1 and the model of RpTRPA5B, shown as sticks.