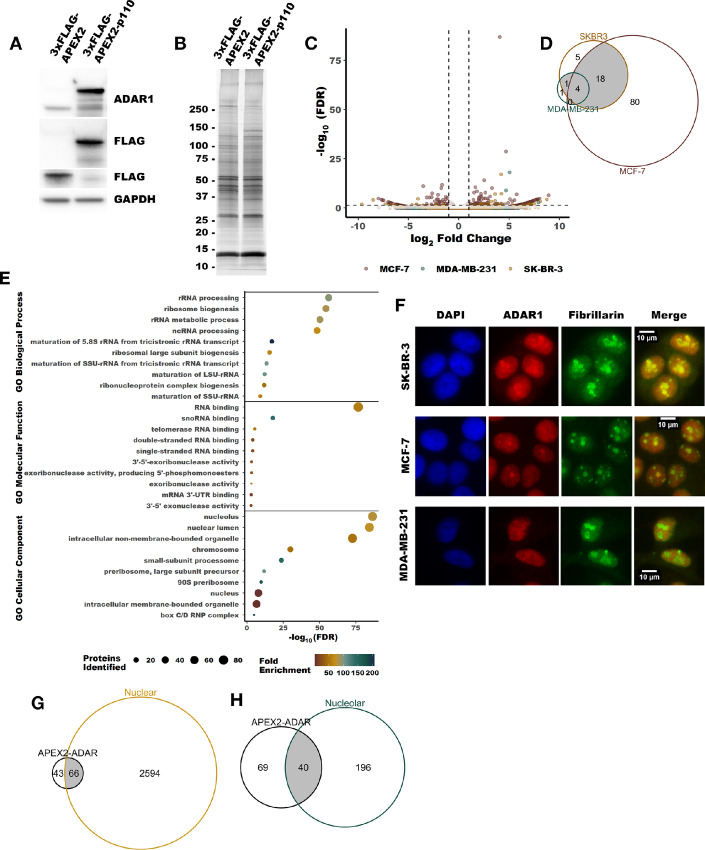
FIGURE 1.
Identification of putative ADAR1-interacting proteins by APEX2 proximity labeling. A, Representative immunoblot showing expression of the constructs used for proximity labeling in MCF-7, MDA-MB-231 and SK-BR-3, MDA-MB-231 is shown. B, Representative fluorescently stained gel image showing proteins purified by streptavidin-biotin pulldown following proximity labeling in MCF-7. C, Volcano plot summarizing the proteins identified by MS following streptavidin pulldown subsequent to proximity labeling. Differential abundance of all proteins in each cell line can be found in Supplementary Table S2. D, Venn diagram showing overlap between enriched proteins from all three cell lines. The cutoff for enriched proteins was an FDR-adjusted P value of less than 0.05 and a log2 fold change of greater than 0.5. E and F, Venn diagrams showing overlap between the enriched proteins identified and those proteins found previously to localize to the nucleus or nucleolus in MCF-7 (57). G, GO terms found to be overrepresented in the list of the enriched proteins. Only the top 10 GO terms, by FDR, are shown for each category. All other significant GO terms are available in the Supplementary Tables S3 and S4. H, Representative indirect immunofluorescence micrographs showing localization of ADAR1, fibrillarin (a nucleolar marker) and DAPI.