FIGURE 4.
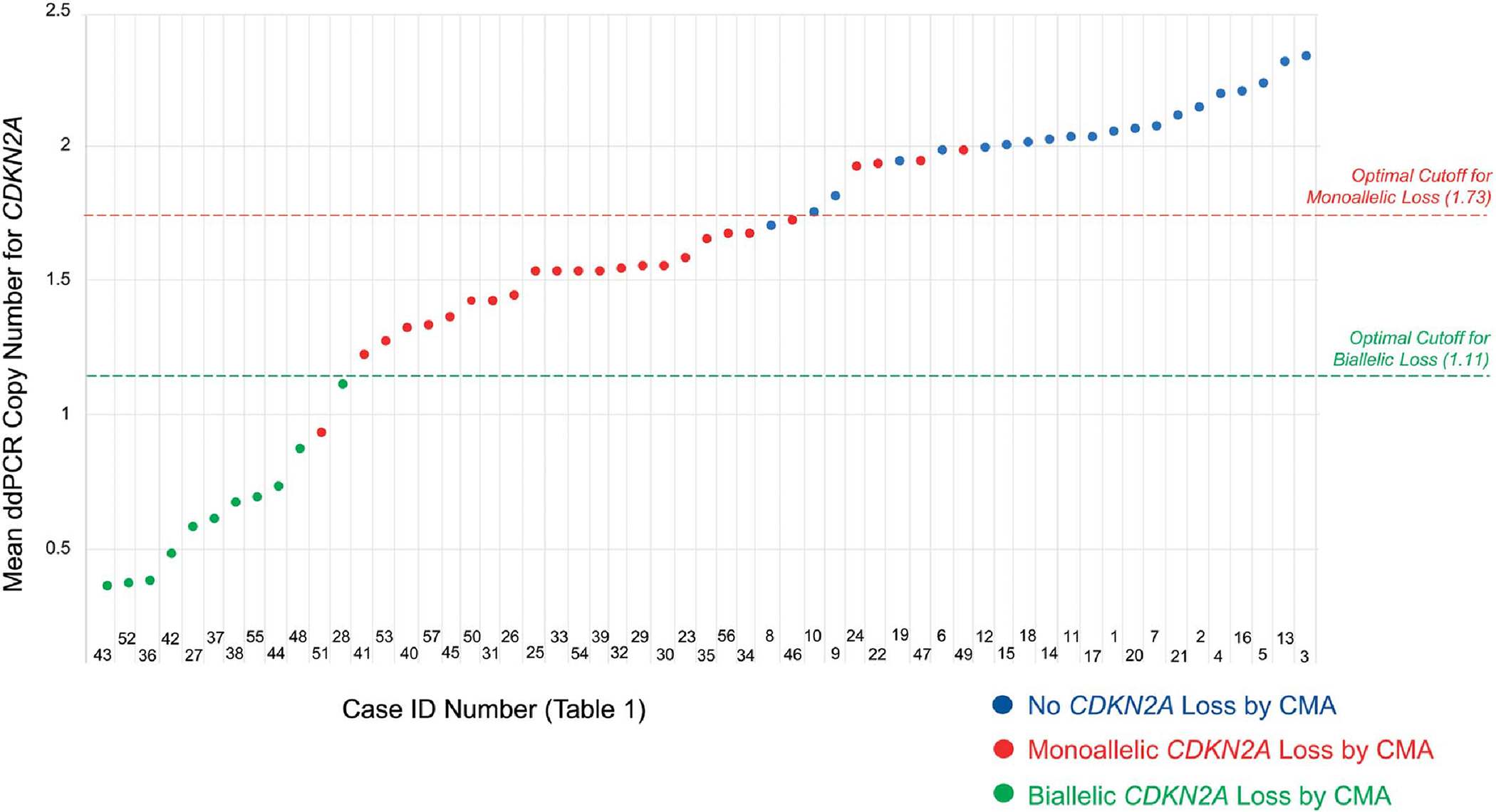
Diagrammatic representation of all cases and mean ddPCR outputs, color-coded by CMA status for CDKN2A. Below a mean ddPCR output of 1.70, all cases have loss of CDKN2A by CMA. Above 2.00, no cases have loss of CDKN2A. Future studies could investigate whether the values in the 1.70–2.00 range reflect lesions with partial malignant transformation. The ROC-determined optimal cutoffs were 1.73 for monoallelic CDKN2A loss and 1.11 for biallelic CDKN2A loss (mean ddPCR >1.73 = no CDKN2A loss; 1.11 < mean ddPCR ≤1.73 = monoallelic CDKN2A loss; mean ddPCR ≤1.11 = biallelic CDKN2A loss).
