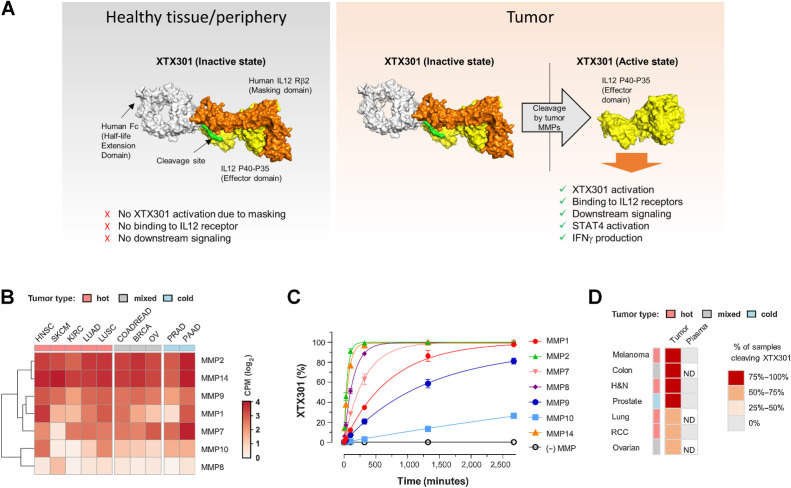
Figure 1.
Multiple MMPs are expressed in both hot and cold tumors and can activate XTX301. A, A schematic of inactive and active XTX301. B, Heat map showing RNA expression levels of genes encoding for MMPs (MMP-1, -2, -7, -8, -9, -10, and -14) in tumor samples from multiple The Cancer Genome Atlas (TCGA) indications (x-axis). Color intensity indicates median log2-transformed CPM reads for each gene and indication. Column keys (pink, gray, blue) indicate prevalently hot (pink), cold (blue) or mixed tumor indications. Abbreviations: HNSC, head and neck squamous cell carcinoma (n = 522); SKCM, skin cutaneous melanoma (n = 472); KIRC, kidney renal clear cell carcinoma (n = 534); LUAD, lung adenocarcinoma (n = 517); LUSC, lung squamous cell carcinoma (n = 501); COADREAD, colon and rectum adenocarcinoma (n = 382); BRCA, breast invasive carcinoma (n = 1100); PRAD, prostate adenocarcinoma (n = 498); OV, ovarian serous cystadenocarcinoma (n = 307); PAAD, pancreatic adenocarcinoma (n = 179). C, Human recombinant MMPs (1, 2, 7, 8, 9, 10, and 14), were evaluated for their ability to proteolytically cleave XTX301 in vitro. The data points represent the mean of 2 independent experiments analyzed as technical replicates (in duplicate) by CE-SDS; the error bars represent the SEM. D, Heat map showing the percentage of human tumors and plasma able to proteolytically activate XTX301. XTX301 was incubated with cells from dissociated primary tumor tissues from melanoma (n = 8, 100% of samples cleaved), colorectal (n = 11, 91% of samples cleaved), head and neck (H&N) (n = 16, 81% of samples cleaved), prostate (n = 21, 76% of samples cleaved), lung (n = 17, 71% of samples cleaved), ovarian (n = 12, 58% of samples cleaved) or RCC (n = 31, 61% of samples cleaved) cancer patients, or plasma from melanoma (n = 5), H&N (n = 10), prostate (n = 7) and RCC (n = 6) cancer patients. Samples were collected after a 24-hour incubation period, and the percentage of cleaved XTX301 was determined by fluorescent triplex western blot. ND, not determined.