Figure 3.
UPR activation and proteostasis during direct neuronal reprogramming of pAstros
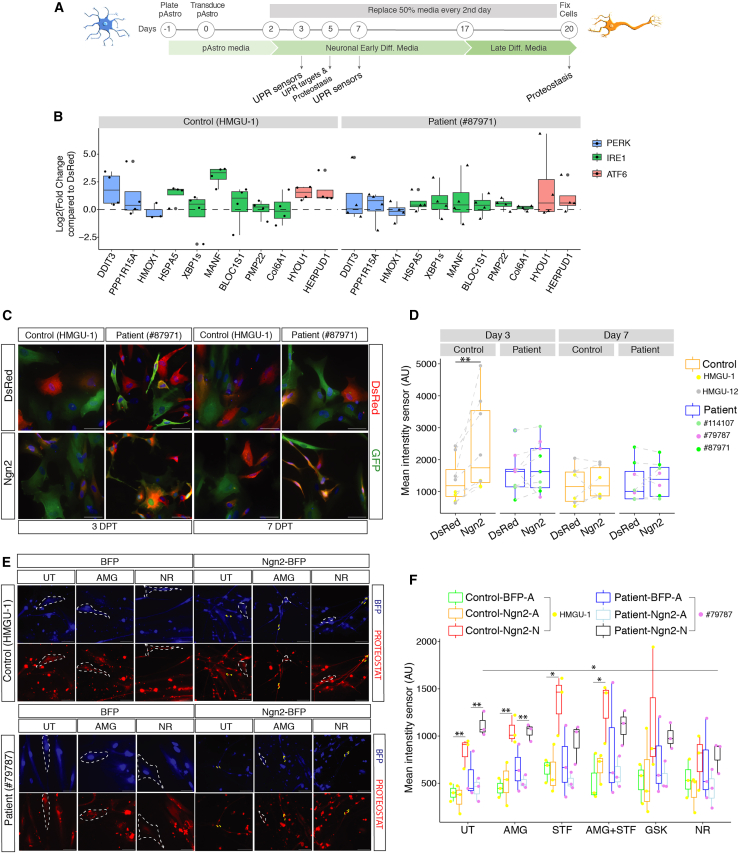
(A) Experimental design.
(B) Boxplots depicting the expression of UPR targets in control (left) and patient (right) pAstros at 5 DPT as measured by RT-qPCR. Data are shown as the fold-change relative to dsRed and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Data are shown as median ± IQR. Each dot represents an independent biological replicate. n = 3–4 independent experiments.
(C) Micrographs of control and patient pAstros transduced with ATF4-YFP sensor and dsRed or Ngn2-dsRed at 3 DPT (left) and 7 DPT (right). Scale bars, 50 μm.
(D) Boxplots depicting mean fluorescent intensity of ATF4-YFP sensor at 3 DPT (left) and 7 DPT (right) in control and patient pAstros. Data are shown as median median ± IQR. n = 3–5 independent batch cultures (2 lines per control and 3 lines per patient). ∗∗p ≤ 0.01.
(E) Micrographs of control and patient pAstros transduced with BFP or Ngn2-BFP with aggresomes labeled with PROTEOSTAT dye at 20 DPT. Scale bars, 50 μm.
(F) Boxplots depicting aggresome detection in control and patient pAstros in different experimental conditions at 20 DPT. Cells are subdivided into Ngn2-A (astrocyte-like) and Ngn2-N (neuronal). Data are shown as median ± IQR. ∗p ≤ 0.05; ∗∗p ≤ 0.01. n = 3 independent biological replicates per line.