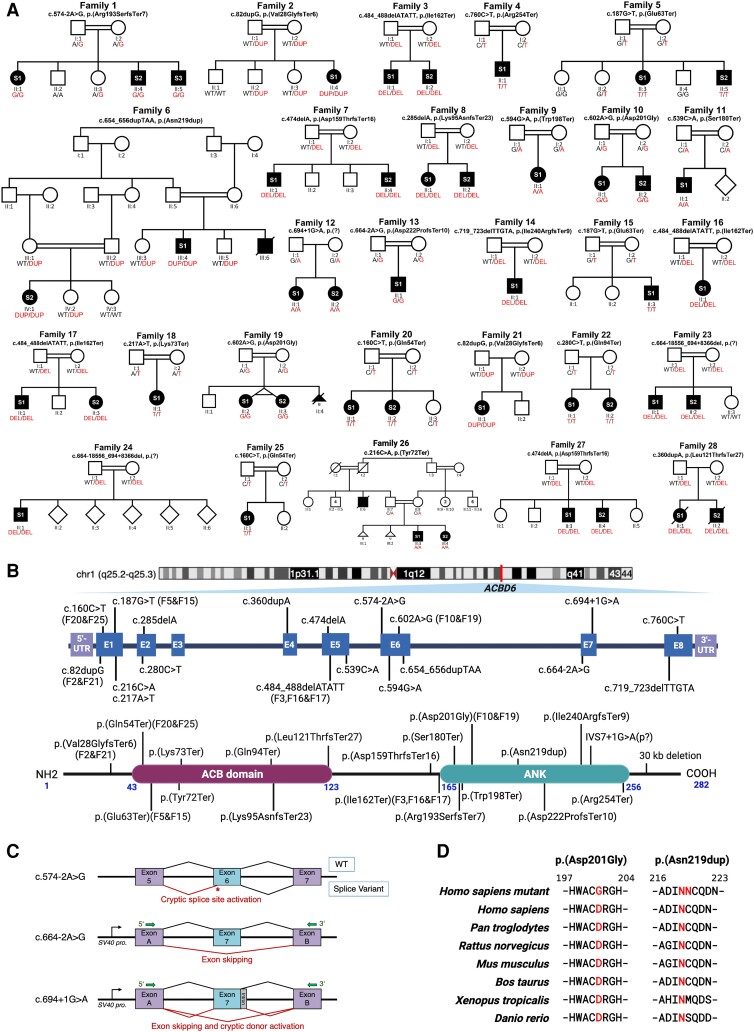
Figure 1.
Family pedigrees, schematic variants’ representation, conserved regions of substitution variants in ACBD6, and splicing effects. (A) Pedigrees and segregation results for the 28 unrelated families. Double lines between individuals represent consanguinity. The 45 affected individuals recruited for the study are shaded and indicated with their respective subject (S) number (S1, S2 or S3). The segregation data for all individuals tested via Sanger sequencing are shown with the presence of the ACBD6 variant (red) and/or the reference allele (black), two red/black texts indicate a homozygous state and one red + one black text indicates a heterozygous state. The genotyping is based on the coding DNA sequence. DEL = deletion; DUP = duplication; WT = wild-type. (B) Top: Schematic representation of the gene and protein positions of detected variants in ACBD6. ACBD6 is located on chromosome 1 at cytogenetic position q25.2q25.3. Middle: The genetic variants mapped to the NM_032360.4 transcript of ACBD6. Bottom: ACBD6 variants mapped on the protein level. Three variants, including p.(Gly22fs), p.(Leu121ThrfsTer27) and a 30 kb deletion in the C-terminus, have been reported previously.6,7,10 Recurrent variants are labelled with family codes. (C) Splicing schematic for the c.574-2A>G (top) variant in ACBD6 showing cryptic acceptor splice site activation in exon 6. The c.664-2A>G (middle) and c.694+1G>A (bottom) variants affect splicing of exon 7 both show exon skipping. Additionally, the c.694+1G>A variant activates a cryptic donor splice site. (D) Interspecies alignment performed with Clustal Omega showing the complete conservation down to invertebrates of the amino acid residues affected by a missense variant leading to an amino acid substitution p.(Asp201Gly) and an in-frame duplication p.(Asn219dup).