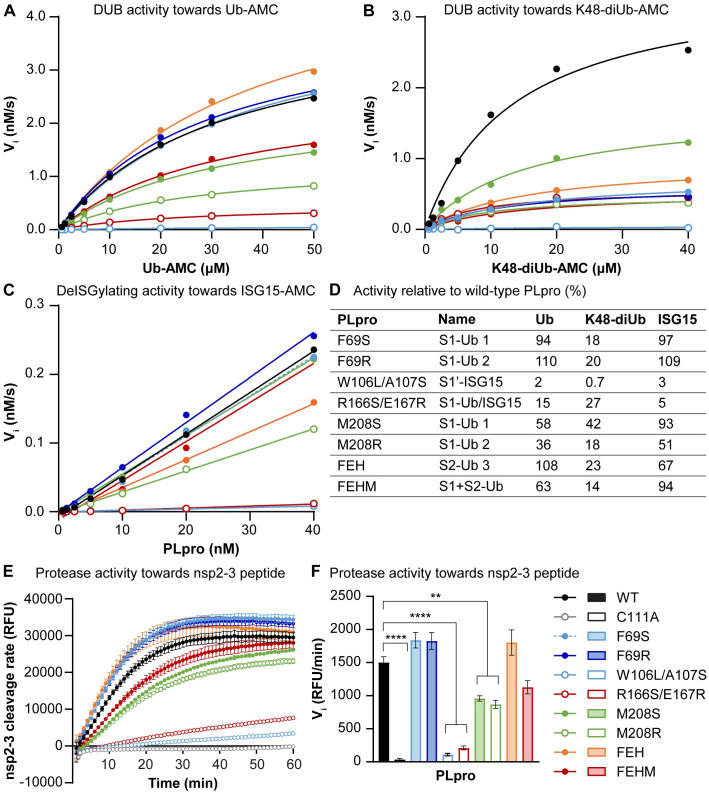
Fig 2. DUB, deISGylating and polyprotein processing activity of SARS-CoV-2 PLpro mutants.
(A-B) Michaelis-Menten kinetics comparing hydrolysis of a range of Ub-AMC (A) and K48-diUb-AMC (B) concentrations by SARS-CoV-2 wild-type PLpro and mutants. (C) Kinetics of a range of SARS-CoV-2 PLpro concentrations towards ISG15-AMC. (D) Activity of PLpro mutants on Ub-AMC, K48-diUb-AMC, and ISG15-AMC relative to wild-type PLpro, as calculated from the kcat/KM values for Ub-AMC and K48-diUb-AMC. For ISG15-AMC, the slope of the curves plotted in panel C were used to calculate relative activity. Naming of mutants corresponds to Fig 1. (E) Polyprotein processing activity of SARS-CoV-2 wild-type PLpro and mutants as assessed by cleavage of a FRET peptide corresponding to the nsp2-3 junction. (F) Initial velocities were extrapolated from the linear portion of the curve as a measure for the substrate cleavage rate. FEH(M) = F69S/E70K/H73G(/M208S) Representative examples are shown of one to three experiments each performed in technical triplicate (A-D). Data in E-F is represented as mean ± s.e.m. of 3 experiments and was analyzed by one-way ANOVA with Dunnett’s multiple comparisons test, comparing each group to wild-type PLpro. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.