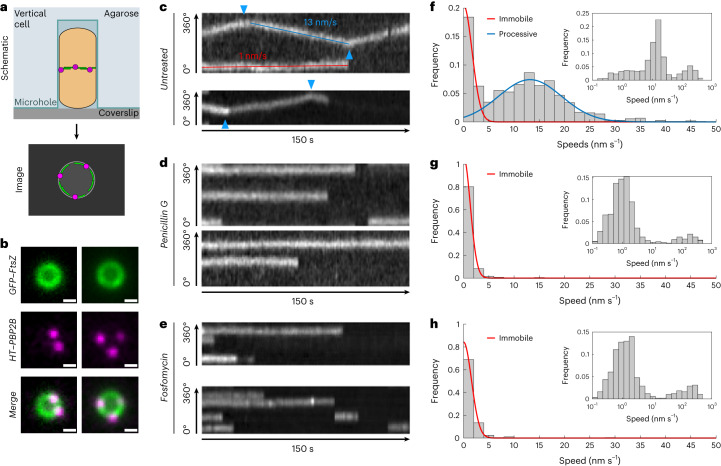
Fig. 1. Two-colour smVerCINI imaging shows a single processive population of HT–PBP2B that is associated with sPG synthesis.
a, Schematic of two-colour smVerCINI experimental setup. A B. subtilis cell (orange) is oriented vertically in a microhole made of agarose (light blue), adjacent to a microscope coverslip (grey). By orienting the cell vertically, the division proteins FtsZ (green) and PBP2B (magenta) can be imaged in a single microscope imaging plane. b, Example images of two vertically trapped cells. GFP–FtsZ (green) is expressed at a low level (0.075% xylose induction) for use as a septal marker, while HT–PBP2B (100 μM IPTG induction) is labelled substoichiometrically with JFX554 HT ligand17 (250 pM) so that single molecules can be easily observed. Scale bars, 500 nm. c, Example radial kymographs of HT–PBP2B motion from smVerCINI videos in untreated cells. Diagonal lines result from processive motion (example shown with blue line), while horizontal lines result from lack of motion (example shown with red line). Blue arrows indicate where HT–PBP2B molecules changed direction. d,e, Example radial kymographs of HT–PBP2B motion from smVerCINI videos in cells treated with 20 µg ml−1 penicillin G (d) or 500 µg ml−1 fosfomycin (e). f, Histogram of HT–PBP2B speeds in untreated cells. Red and blue lines show fits to the data (Methods). Inset: histogram of speeds plotted on logarithmic x axis, showing three populations. g,h, Histograms of speeds in cells treated with penicillin G (g) or fosfomycin (h). Red lines show fits to the data (Methods). Insets: histograms of speeds plotted on logarithmic x axis, showing two populations. All examples in c–h are from cells grown in rich media at 30 °C. Sample sizes are listed in Supplementary Table 6.