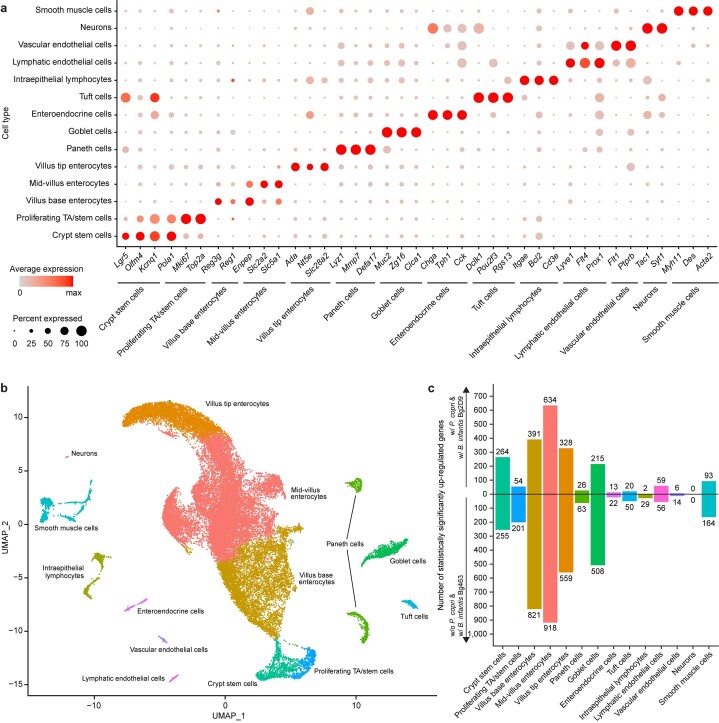
Extended Data Fig. 5. snRNA-Seq analysis of differential intestinal gene expression in mice colonized with bacterial consortia with or without P. copri.
Jejunal tissue samples collected from ‘w/ P. copri & w/ B. infantis Bg2D9’ and ‘w/o P. copri & w/ B. infantis Bg463’ groups at the end of the experiment described in Fig. 1 were analyzed. (a) Dot plot of marker gene expression across epithelial cell types. The average expression level and percentage of nuclei that express a given gene within a cell type are indicated by dot color and size, respectively. (b) Integrated UMAP plot for all jejunal nuclei isolated from animals in both arms (n = 4 mice/arm). (c) The number and directionality of statistically significant differentially expressed genes in each cell cluster.