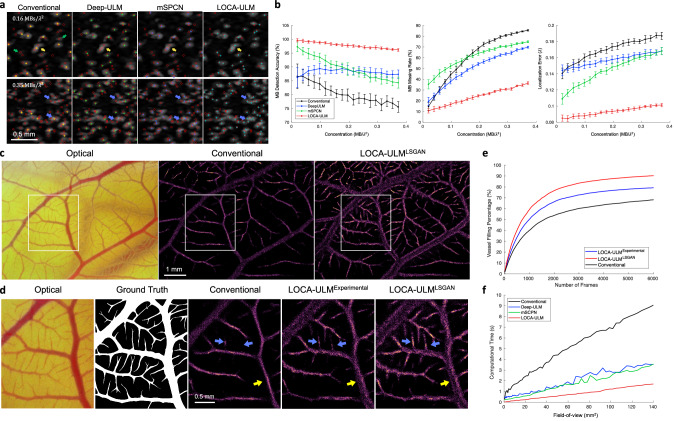
Fig. 2. Results of the simulation study and in vivo chicken embryo CAM imaging study.
a Simulation results of conventional localization, Deep-ULM14, mSPCN21, and LOCA-ULM with low () and high () MB concentrations. Ground truth MB positions are marked by red circles, conventional localization by yellow , Deep-ULM by green , mSPCN by blue , and LOCA-ULM by cyan . b Comparison between conventional localization, Deep-ULM, mSPCN, and LOCA-ULM was performed on simulated frames at increasing MB concentrations, using three performance metrics: MB detection accuracy, MB missing rate, and localization error. The error bars indicate the standard deviation of inference over 500 frames. c–e Comparisons among conventional localization, LOCA-ULMExperimental, and LOCA-ULMLSGAN MB localization in in vivo CAM imaging. c Optical microscopy image of the CAM surface microvessel, along with MB localization images reconstructed by conventional localization and LOCA-ULMLSGAN. n = 1 experiment. d The ROI selected from the optical image and the corresponding ground truth vessel segmentation. Magnified view of the MB localization images marked by the white ROI for conventional localization, LOCA-ULMExperimental, and LOCA-ULMLSGAN. e The vessel filling (VF) a percentage of conventional localization, LOCA-ULMExperimental, and LOCA-ULMLSGAN, as a function of the number of frames (Methods). f Comparison of computational times for MB localization across varying FOV sizes using conventional ULM, Deep-ULM, mSPCN, and LOCA-ULM. Computational times encompass the MB localization process, which includes normalized cross-correlation and regional maximum search for conventional ULM, a network forward pass for LOCA-ULM, and a network forward pass with centroid calculation for both Deep-ULM and mSPCN. Source data are provided as a Source Data file.