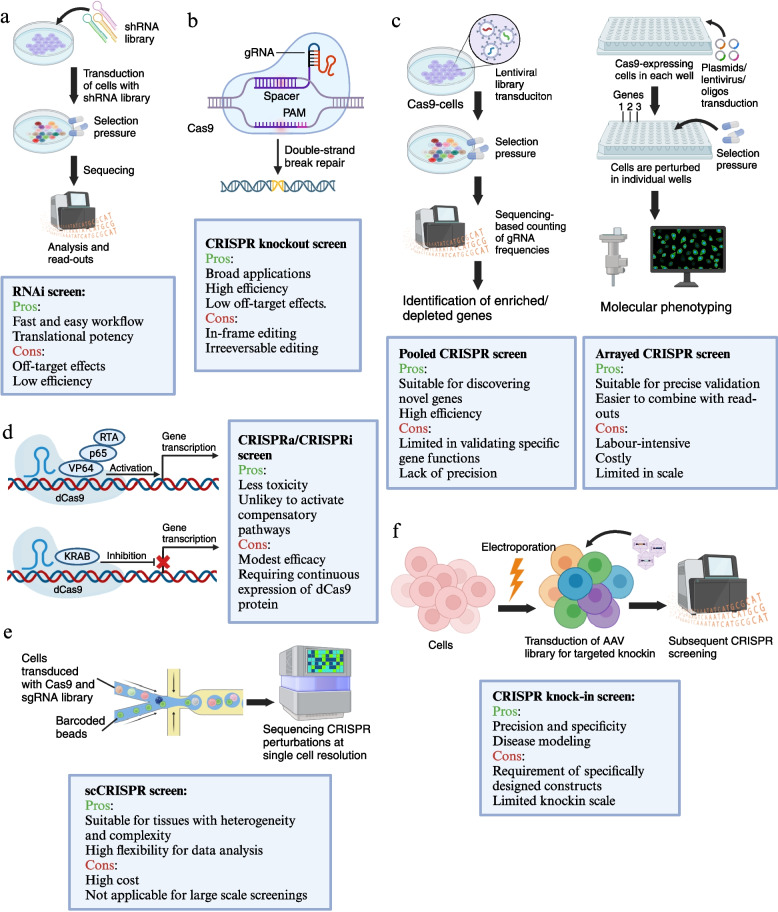
Fig. 3.
Different CRISPR screening methods are described with their applications, pros and cons. (a) RNAi screen. In RNAi screens, RNAi reagents were introduced into the cells to target the endogenous mRNA transcripts. By combining RNAi tools with analysis such as signal transduction, cell viability, and responses to infections, RNAi screening enables identification of new genes, and the information of gene function in a wide variety of biological processes. (b) CRISPR knockout screen. Directed by a guide RNA (gRNA), Cas9 nucleases introduce double-strand breaks (DSB) into the target site; subsequent DNA repair results in compromised gene function. (c) Pooled/arrayed CRISPR screen. Pooled CRISPR screens introduce perturbations in bulk, genetically encoding them, and commonly employing gRNA sequencing for readout. Arrayed CRISPR screens involve the separate introduction of distinct perturbations. Since each reaction compartment undergoes a defined perturbation, the read-out does not necessarily require gRNA sequencing. (d) CRISPRa/CRISPRi screen. CRISPR activation (CRISPRa) screen employs dCas9 coupled with transcriptional activators, such as the VP64 domain, resulting in the stimulation of genes in target site. CRISPR interference (CRISPRi) screen uses dCas9, fused with transcriptional repressors like Krüppel-associated box (KRAB). This fusion results in the repression of genes in proximity to the gRNA target site. (e) scCRISPR screen. scCRISPR (single-cell CRISPR) screens combine pooled CRISPR perturbations with scRNA-seq, offering opportunities to investigate genome regulatory networks by interrogation of different perturbations with the transcriptome profiles at single-cell resolution. (f) CRISPR knock-in screen. CRISPR-based knock-in screens mediate simultaneous gene editing and precise transgene knock-in, producing cell pools with targeted stable gene editing