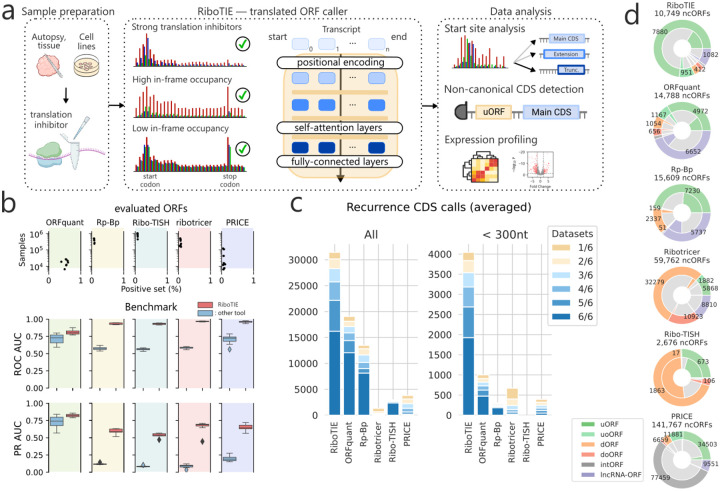
Figure 1: Machine learning to delineate RNA translation from Ribo-Seq data with RiboTIE.
a, Schematic that outlines the flexibility and function of RiboTIE as a machine learning model (transformers) for ribosome profiling data. b, Benchmarking analyses featuring eight datasets. RiboTIE is compared with five other tools for translated ORF delineation from ribosome profiling. Precision recall (PR) or Receiver Operator Characteristic (ROC) Area Under the Curve (AUC) scores are compared on ORF libraries that are unique to each tool. c, A stacked barplot that reflects the number of called annotated CDSs (le, all; right, <300nt) by each tool for six replicate samples of pancreatic progenitor cells, the fraction of CDSs that are found in a certain number of replicates is represented as well. d, The total number of non-canonical ORFs (ncORFs) and each type of ncORFs called by each tool combining all predictions on the six replicate samples of pancreatic progenitor cells. The inner fractions represent ncORFs present in >4 datasets.