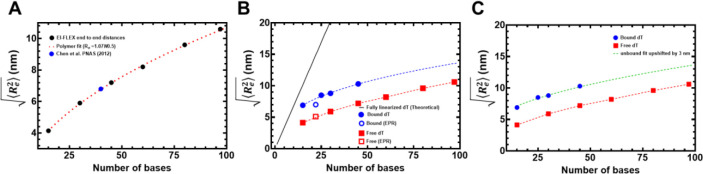
Figure 6. Computational predictions of ssDNA wrapping agree well with experimental measurements.
A) Black squares are data, and the red dashed line is a fit to those data using a polymer scaling model. The blue circle is data reporting on the end-to-end distance for (dT)40 made by Chen et al. 2011). B) Blue circles are from RPA-bound (dT) oligonucleotides, red squares are from RPA-free (dT) oligonucleotides. Data include distances measured by both EPR and smFRET experiments. Blue and red dashed lines are polymer model fits to the data, respectively. The black line is the theoretical expected end-to-end distance if the DNA were fully linearized and stretched to its contour length. C) The root mean squared end-to-end distance dependence on the number of bases is essentially identical for the bound and unbound states, as highlighted by the fact that the exact polymer fit upshifted by ~3 nm fully describes the bound dT dependency.