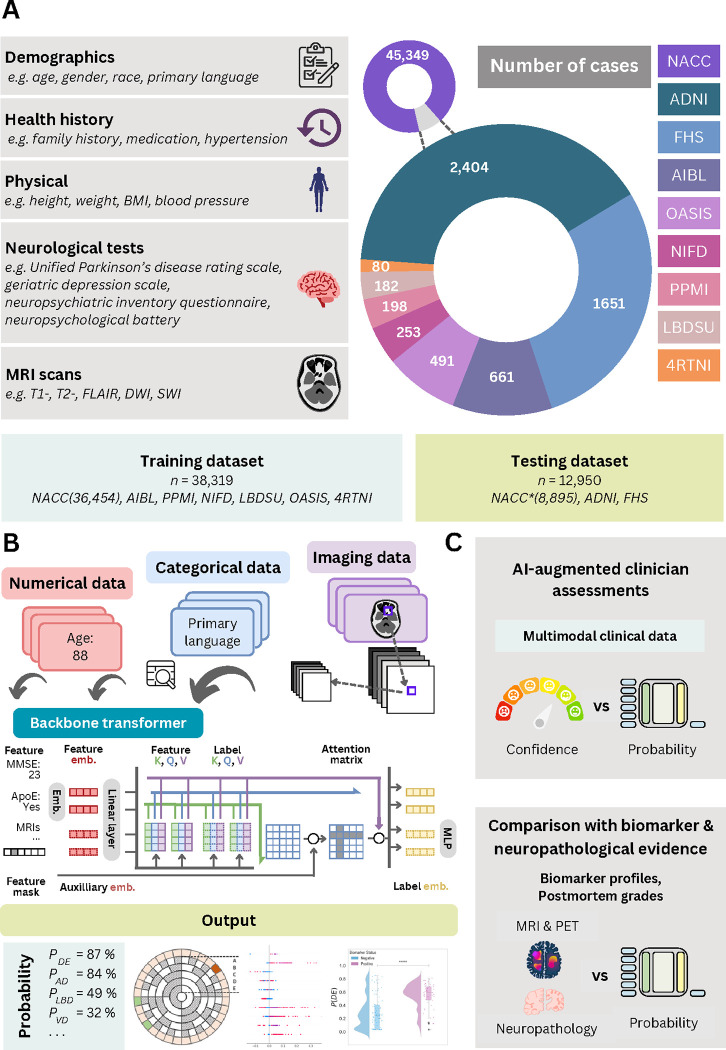
Figure 1: Data, model architecture and modeling strategy.
(a) Our model for differential dementia diagnosis was developed using diverse data modalities, including individual-level demographics, health history, neurological testing, physical/neurological exams, and multi-sequence MRI scans. These data sources whenever available were aggregated from nine independent cohorts: 4RTNI, ADNI, AIBL, FHS, LBDSU, NACC, NIFD, OASIS, and PPMI (Tables 1 & S1). For model training, we merged data from NACC, AIBL, PPMI, NIFD, LBDSU, OASIS and 4RTNI. We employed a subset of the NACC dataset for internal testing. For external validation, we utilized the ADNI and FHS cohorts. (b) A transformer served as the scaffold for the model. Each feature was processed into a fixed-length vector using a modality-specific embedding strategy and fed into the transformer as input. A linear layer was used to connect the transformer with the output prediction layer. (c) A subset of the NACC dataset was randomly chosen to conduct a comparative analysis between neurologists’ performance augmented with the AI model and their performance without AI assistance. Similarly, we carried out comparative evaluations with practicing neuroradiologists, who were provided with a randomly selected sample of confirmed dementia cases from the NACC testing cohort, to assess the impact of AI augmentation on their diagnostic performance. For both these evaluations, the model and clinicians had access to the same set of multimodal data. Finally, we assessed the model’s predictions by comparing them with biomarker profiles and pathology grades available from the NACC, ADNI, and FHS cohorts.