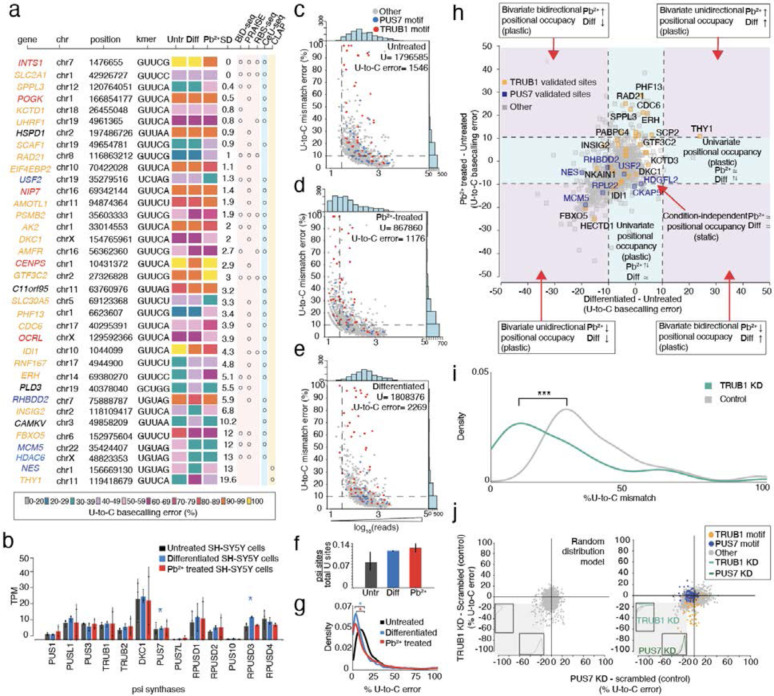
Figure 3. Psi analysis across three cellular states enables the classification of plastic and static sites of modification, transcriptome wide.
a. Heatmap of sites with at least 40% U-to-C basecalling error in one of three conditions. Colors indicate the percentage of U-to-C basecalling errors. For each position the standard deviation (SD) is calculated on all three conditions and used to rank the sites in order, with the most similar at the top and the least similar at the bottom. KD validated substrates for TRUB1 are shown in orange, and TRUB1 motifs that have not yet been validated are shown in red. KD validated substrates for PUS7 are shown in dark blue, and PUS7 motifs that have not been validated are shown in light blue. Orthogonal validation with non-nanopore methods is performed for all the sites.
b. DRS determined the TPM of various PUS enzymes in untreated, differentiated, and Pb2+-treated SH-SY5Y cells. Individual colored bars represent each experimental condition, with error bars describing the standard error of the mean (SEM) across downsampled replicates. Individual replicates are shown as black dots. Statistics are performed by Student's t-test, comparing each KD group to the scrambled control sample. * p < 0.05, ** p < 0.01, *** p < 0.001.
c. Untreated SH-SY5Y number of total DRS reads are plotted against the U-to-C mismatch error of putative Ψ sites detected by Mod-p ID (p-value < 0.001 in at least two biological replicates). Blue dots represent uridine positions within a PUS7 motif, while red dots represent uridine positions within a TRUB1 motif. All the other motifs are shown in grey. Dashed lines represent the criteria used for defining a position as a psi site (number of reads>10; U-to-C error>10).
d. Pb2+ treated SH-SY5Y number of total DRS reads are plotted against the U-to-C mismatch error of putative Ψ sites detected by Mod-p ID (p-value < 0.001 in at least two biological replicates).
e. Differentiated SH-SY5Y number of total DRS reads are plotted against the U-to-C mismatch error of putative Ψ sites detected by Mod-p ID (p-value < 0.001 in at least two biological replicates).
f. Colored bars shows the number of significant pseudouridine sites, labeled as U-to-C error in panels d,e,f, normalized by the total number of reads in each of the three conditions. Error bars show the standard error of the mean across biological replicates.
g. Distribution of the occupancy levels calculated on untreated, differentiated, and lead-treated libraries. Statistical significance is calculated using 95% CI, * p < 0.05.
h. Putative psi-positions determined by Mod-p ID are plotted according to the difference in U-to-C basecalling error in the untreated and Pb2+ treated samples against the difference in U-to-C basecalling error between the differentiated and untreated for each position. A dotted line at the +10% and −10% marks indicate the cutoff for a position to be changed in response to perturbation. Condition-independent positional occupancy (static) sites are shown in the center square. Single condition-dependent positional occupancy sites are shown in the blue stripes. Double condition-dependent positional occupancy sites are shown in the purple areas.
i. Distribution of the occupancy levels calculated on PUS7 validated sites before (grey) and after the TRUB1 KD (green). The occupancy levels in the KD library are decreased compared to the control condition without KD. Statistics are performed by Student's t-test, comparing each KD group to the scrambled control sample. * p < 0.05, ** p < 0.01, *** p < 0.001.
j.Co-operation effect of TRUB1 and PUS7 KD on SH-SY5Y cells. (Left) Random distribution model of points constrained by the data parameters. Distributions are calculated for points in the lower left quadrant as a function of each simulated knockdown. (Right) distribution of TRUB1 KD plotted against PUS7 knockdown. Distributions are calculated for points in the lower left quadrant as a function of each knockdown.