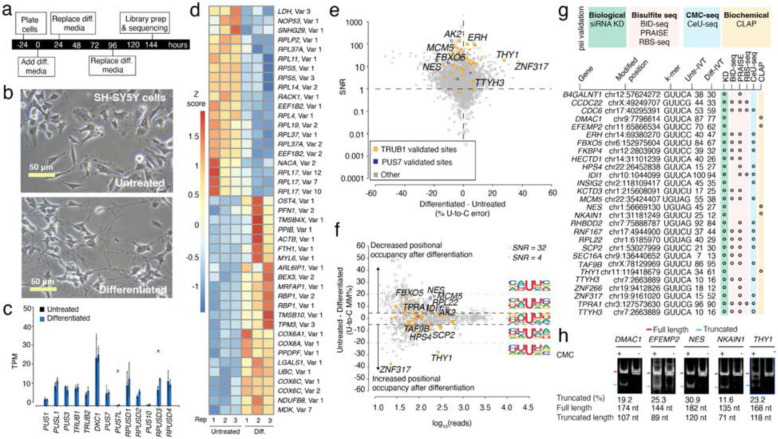
Figure 1. Effects of RA-mediated differentiation on mRNA psi modification and machinery in SH-SY5Y cells.
a. Timeline illustrating the stages and duration of the RA treatment applied to SH-SY5Y cells.
b. A representative photomicrograph of untreated and differentiated SH-SY5Y cells is shown.
c. TPM of various PUS enzymes in untreated and differentiated SH-SY5Y cells determined by DRS. Individual colored bars represent each experimental condition, with error bars describing the standard error of the mean (SEM) across downsampled replicates. Individual replicates are shown as black dots. Statistics are performed by Student's t-test, comparing each KD group to the scrambled control sample. * p < 0.05, ** p < 0.01, *** p < 0.001.
d. We used Deseq2 to identify the transcripts with the highest fold change between the untreated and differentiated samples. Three biological replicates for each condition were used. The color scale shows a Z-score based on the relative fold-change.
e. SNR vs. the difference in U-to-C error % between the untreated and differentiated samples. Orange dots represent uridine positions that are validated TRUB1 substrates, and blue dots represent uridine positions that are validated PUS7 substrates.
f. Putative psi-positions determined by Mod-p ID are plotted according to the difference in U-to-C basecalling error in the untreated and differentiated samples against the reads for each position. Dotted line at the +5% and −5% marks indicate the cutoff for a position to be changed in response to perturbation. Inset shows the sequencing logo for positions within the TRUB1 motif, gray points above the threshold line, and total points above the threshold line.
g. Annotation of genes containing a psi modification validated by PUS7 or TRUB1 KD (Figure 2) and orthogonal methods.
h. CLAP gel result of psi incorporation in mRNAs for the sites confirmed by Nanopore DRS and KD data, but not by any other orthogonal method. See Supplementary Fig. 2 for original gel source data.